Hi, this is a comment.
To get started with moderating, editing, and deleting comments, please visit the Comments screen in the dashboard.
Commenter avatars come from Gravatar.
For what I understood V.harveyi can be considered a “cheater” because it encodes genes for the byosinthesis of amphi-enterobactin which is has a high affinity to siderophore that acquires iron formicrobial systems.
Interesting experiment, it explained very well how the aerobactin works as a inhibitor molecule for V. harveyi and taking in count that this lead to a quantitative investigation the results can be more precise in future experiments.
After reading about FISH I was impressed with the many adventages it has, since it allows the detection of one to three orders of magnitude more bacterial cells in the samples, allows study of the spatial organization of cells and in top of that, cells do not have to be alive.
I was also wondering the same thing about the use of icebergs to supply cities with water and it could bring more damage to the environment than it seems. The towing of icebergs can cause a change in water temperatures which could affect the aquatic life of the area creating a chain reaction that would distort the ecosystem of Antarctica.
Taking into account that the alternative of using icebergs as a source of water supply is not very viable due to the alteration that it would cause in the ecosystem, it is possible that these contaminated icebergs can be treated after bioremediation for an emergency case due to Lack of water ?, Is this “fresh water” still drinkable?
I had the same question and I have done some research. It seems that over long periods of time (two years) it can have an impact on the PH levels causing them to decrease. However, taking in count that the waiting time it was not so long it would not make much of a difference. But, I’m still not so sure why they waited two months to do the analysis.
For what I researched the agitation moves the liquid onto the walls of a vessel and it can increase the surface area which would subsequently increase the oxygen concentration allowing the bacteria to grow faster.
Situ bioremediation caught my attention and it seems that there are many methods that can be used. One of them is the biosparging that involves the injection of air under pressure below the water to increase water oxygen concentrations.
yeah that caught my attention too. Foreign organism are forbidden because of the isolates location, meaning that the local organism have not developed natural protection against foreign species.
When they say “we restricted our analyses to their free-living lifestyles using shaking liquid culture for co-incubations”, what kind of specific culture do they mean?.
It is really interesting how this species interact with each other and the fact that V. Fischeri ES114 can use a filtration method of a 10000 MWCO membrane to prevent V. Harveyi from growing.
As stated in the introduction, nitrification only have a good performance in the lower oxic zone, this is because one of the main factors that nitrification needs its a good concentration of oxygen and as we go deeper in water this concentrations of oxygens star to decrease making it very difficult to carry out the process in the suboxic and anoxic zones.
(A). the crenarchea cells showed better performance in the suboxic area and very low or non existent performance in the lower oxic and anoxic areas.
(B). BAOB cells and mRNA performed very poorly inall the zones.
(C). YAOB cells in the suboxic zone performed their best during this time but their performance was very low in the lower oxic and anoxic zones. However, mRNA did not have fized performance where it decreased and increased between zones.
Yes, because port activies can provoke a lot of pollution with ships, transport, diesel emision, and loading of products. Also, we can add the risk of a possible oil spill. In conclusion, Mobile bay could have greater pollution due to high activities than other places that are not this active.
Good point, MSM would be a good media to start specially because of what the scientist are looking for which is a bacteria that can tolerate high concentration of salt like the ocean. After, we can exclude those who can’t biodegrade the hydrocarbons in a high salinity enviroment.
MSM media is considered a slective and differential growth media at the same time because it encourages the growth of certainbacteria while decreasing the growth of other kind of bacteria. In the other hand, we have Lurie Broth is a rich mediaum that allows the growth of a different bacteria and it consist of tryptone, yeast and sodium chloride. The reason that they used both its because by having a rich medium it is highly probable that it would grow in that medium but in the MSM there could an exclusion.
I did some investigation about the UltraClean soil DNA kit and I found it that the soil samples are added to a bead beating tube containing beads, lysis solution, bead solution and inhibitor removal solution. The obective is to lyse the microorganisms in the soil by a combination of heat, detergent, and mechanical force against specialized beads.
it is really interesting that seminole was able to use benzene as a carbon and energy source but the situation changed when the bacteria could not dregrade benze at a 4 M NaCl.
I was curious about this topic two and I did some research an catechol can be degradaded by two ways: the meta cleavage pathway or the ortho-cleavage pathway.
I look up fluorene and is PAH used to make dyes, plastics and pesticides. Also, one interesting fact that I found is that it has an aromatic odor similar to naphthalene. It is possible that compounds that have similar odors could have similar degrading bacteria, like there could be any correlation?.
interesting that they also have problems identifying the enzymes that govern the initial attack on the PAH degrador as the scientist from the previous paper.
interesting that they are using gas chromatography for the separation of mixtures. we just used column chromatography separation methos which is similar to the gas method, helped me to understand.
I was curious about gene transfer so I ended up google in it and it says that is the movement of genetic material between organisms belonging to distinct groups of interbreeding individuals.
I googled Fluoranthene and is a molecule that can be viewed as the fussion of naphthalene and benzene unit connected by the previously said five-membered ring. However, Fluoranthene is a colorless compound.
I did some research and a study says that the Indian Ocean is the second- most polluted in the world which makes it a escenario to do this kind of research.
I find it so interesting on how the PCR technique works. By just going through cycles of different temperatures resulting the the amplification off DNA is very intriguing.
I believe the environmental factors play a huge effect. Even with the previous paper the bacteria that is closest to that region had the best result of being the most effective in PAH degradation. I am assuming that’s the same case for this paper.
I know we are not supposed to question too much about the details of the technique, but I truly do not understand the treatment of the controls and what is being compared. Or how it contributes to the overall experiment.
I did some research on the neighbor-joining method, because I was curious also. and it does not seem to be the most popular to make phylogenic trees, but it is a quick way to organize a large amount of data. One advantage is that it shows how everything does not evolve at the same rate.
I was curious about the same thing. How is this information will be used to help solve the overall problem? How are they going to take this information and possibly enhance the productivity of the PAH degraders?
It is interesting how there are structural variations of Se(0) particles from different bacteria that are present. I wonder through evolution how that came to be. Like are the systems similar or just the product. Is it due to an ancestor or just environmental pressures over time?
The purpose of adding antibiotics to the media to prevent contamination from other strains. The strains they are trying to isolate are resistant to the antibiotic so in theory its the only strain able to produce colonies.
I believe it is more of a confirmation test. If the bacterial strain is so showing it has the ability to degrade selenium, then there must be a transcriptional pathway that gives the bacteria the tools to degrade.
Yes this appears to be the case, showing that the cosmic clone pECL1e has a significant effect in Se(VI) degradation. This also covers any unforeseen chemical reactions that might have contaminated this test.
That what I go from this paragraph. The purpose was to find out how Se(VI) can be degraded and for this bacterial strain it has to live in anaerobic conditions forcing the strain to use Se(VI) to the the sole electron acceptor this in turns needing FRN regulation to become active.
So the overall result is that the environment greatly dictates the metabolic pathways. It uses “oxygen sensing proteins” in order to to decide which regulatory pathway would be most efficient. So when these proteins do not sense oxygen then that is when the FNR regulator becomes active allowing the bacteria to degrade Se(VI).
I was reading up on the mobile chemistry plant pollution in Chickasaw and I was wondering is it possible that there are cancer possibilities through the aromatics and chemical waste. I’m asking for a family member.
If aerobic non-sporulating rhodococci are great contributors to the biodegradation of environmental pollutants, would it be hazardous to introduce more into the environment to help speed up the end of pollution?
Along with the diluted culture spreads, I was thinking that a concentrated culture could also be spread on a agar plate to compare between the dilute agar plate spreads.
I noticed that the experiment used MSM plates instead of TSA and R2A like we use in our experiments. What exactly are the differences between using MSM plates compared to the others?
If complete degradation of concentrated Fla is the overall goal of the experiment in this section, why not prolong the incubation period? Will it mess up the data?
I’m not quite sure I’m understanding the chart here, but is it basically saying that every relevant match with a percent identity of 86 shows a closer homology to Rhodococcus sp. CMGCZ than the relevant matches that show a percent identity of 84 and 82?
So I’m assuming that LMW PAH has 2-3 ring structures while HMW PAH has 4-5 ring structures. I find it interesting that even though LMW PAH is more favorable than HMW PAH when it comes to degradation, some bacterial strains degrade HMW more readily than LMW, despite ring structure size.
If Nap, Phe and Fla degradation was inhibited on a Ye medium plate, would it have done the same thing if Nap, Phe and Fla were added on a R2A, TSA or MSM plate?
I aslo think that marine bacteria are able to degrade PAH more readily because the environment they live in might be a little bit harsher compared to terrestrial bacteria, for example, saltwater compared to a forest.
I’m also interested in how terrestrial and marine bacteria differentiate between each other when it comes to degrading PAH. If environment plays such a key role in how quickly they degrade it, then we could possibly colonize and utilize them in quicker degradation of harmful substances.
I did not know that PAH affected the surrounding environments and people that live in them to that extent. I think this is very useful in bringing awareness to the situation so ideas can be brainstormed to fix and prevent this problem.
I love how we are able to do these experiments in our lab to see how we could apply this to real world problems. Even though these experiments are a little more in depth than ours, I still enjoy learning and applying techniques to these types of problems.
For the thin layer chromatography, is there a reason why silica gel is used? I’m just curious because i know that you could also use cellulose and aluminum.
Hey Jennifre. I had the same exact question that you stated. Maybe it could be due to some gene silencing or maybe it’s just expensive to keep doing multiple cloning.Not entirely sure though. Just throwing out some suggestions.
Could the reason why the cis-dihydrodiols of the latter compounds were converted to monohydroxylated compounds due to ring stability and aromaticity? Also, could it be due to activating and deactivating groups that are present in the ting structures or the types of reagents used on them that made them become monohydroxylated?
Isn’t upper pathways how the original compound goes to a intermediate compound that still contains ring structures, but they are unstable and lower pathways are rings that are being cleaved and de-aromatized?
Is the reason why the data for the location of the genes encoding PAH dioxygenase of A5 wasn’t shown because it was irrelevant to the paper or was it too much to be included or too complicated?
I was thinking the same thing Tim. This makes me wonder if there are other chemolithotrophs and cyanobacteria that share a symbiotic relationship to help each other and do the same thing when it comes to breaking down arsenic to use for energy.
I wonder if it had mattered whether they chose from these springs or other springs that are around the area if they all contained dissolved sulfide, ammonia, methane and arsenite.
This is very interesting to me. I wonder if they would have the same results that they obtained from this same location either now or last year and whether different steps could be taken with the new technology they possibly have now.
I was wondering the same thing Sarah. To me it would make since that it would find an alternative way to survive if exposed to a darker region than what is was previously in. If this is true, then this is a cool way to examine how adaptation to a new environment plays a role in metabolic pathways.
Hey Lameace, I’m curious as well of whether there is more than one clone and whether this experiment was successful or not. Maybe they could specify that there are more (or just one), but because this was the more dominant arsenic cycler, they concentrated on it more for the sake of the paper.
I was wondering the same thing Jessica. If a sample was taken from a lake that contained less salt than the lake they obtained the biofilm from, would there be a drastic change or a slight change. This is very interesting to me.
Could the temperature ranges from the experiments be due to the environment that they came from (the springs being around 45C and the microbials halting activity at around 50C)?
I agree with your question Jessica. If they already knew that sulfide and natural gases help to drive the reduction to As(V), then why was the experiments incubated with the same electron donors. Maybe they could’ve used a different electron donor to observe the reaction.
If acetate did not give the needed results for the experiment then what did it do? i think that they should include the results of what was observed with the experiment cause I am actually curious as to what happened.
I’ve always associated aromatics with being more good than harmful. I guess because of ‘aroma’ being apart of the name. I knew that their were some that could do some damage, but to the extent that it has caused in The Persian Gulf.
Is there any particular scientific reason why sediment samples were taken from 12 cm below the surface and seawater samples taken from a depth of 15 cm?
Since the strain was found degrade several other aromatic compounds, is this indicative of phenanthrene being more stable than naphthalene?
Because of the ability to degrade more than one compound would phenanthrene be a better aromatic to use concerning the issue of oil contamination in the gulf?
My species from 314 was Burkholderia (Paraburkholderia, specifically). When I was researching it, what I found concerning PAH degradation only spoke of toluene. It interesting to read about the study of another of its degraders.
The fact that this strain was the first fluoranthene-degrading bacterium to be found with the family of Rhodobacteraceae just goes to show how much of the world had yet to be researched, studied, and discovered.
What is the first discovered PAH? I’m sure it has been widely studied. Would that not be a good basis for studying the physiology of not just fluoranthene, but other PAHs as well?
Since xylE and ahdA1b-1 had consistent results to each other when promoted by Cu(II), would this mean that both genes have similar phylogeny or genetic makeup?
Why is there not a lot of data concerning fluorene and gram-negative bacteria? Does that mean gram-positive is more associated with this particular PAH?
Is the reason that phenanthrene cannot be used as the sole carbon source because when the strain is in the presence of another substrate it uses up that substrate first? I may be completely off with my reasoning here.
Even though it was weakly detected, doesn’t that still mean that the strain was able to used glucose? Or would you just ignore that data because it wasn’t substantial enough?
The other papers never mentioned a half-life, if I recall correctly. Does this make carbofuran significantly different or do all PAHs have similar half-lives?
Being that the pink/light red color was more intense in the medium without additional nitrogen source, this basically means that when carbofuran is able to take up the nitrogen is changes in color similar to other PAHs turning blue/black?
I had this same question upon reading this section. Another possibility is a novel gain-of-function mutation, with analogous protein function to other extant PAH pathways in terrestrial bacteria. I also wonder if the ‘acquisition’ could be an activation of a pathway that may be just inactive in other marine bacteria? It could mean a lot of things, and makes me want to read the cited paper.
It seems like there are going to be a lot of results to read, as indicated by this statement. I am most interested to see a comparison of similarities and dissimilarities between the C. indicus PAH degradation pathway and those characterized terrestrial bacteria. Do they involve protein homologs? Do they produce the same intermediates and ultimate products? Are the products of the degradation waste products, or metabolites, or otherwise useful molecules for the organism? What is the energy investment into the degradation? Will the organism maintain the function of the pathway under stress? I hope to see the answers to some of these questions in the results.
Hi Annie,
From what we know about PAH degradation so far, I think it requires oxygen, because the enzyme RDH needs molecular O2 to break the aromaticity of one of the rings and start the process. There may be another enzyme that can do the same thing without O2 though, that would be interesting to learn about for the sake of comparison.
I don’t want this to count for one of my comments, but how cool would it be to get to go diving or in a submersible to collect sediment from the ocean floor for your project? That’s peak biology right there.
Does the term ‘metabolites’ refer to the products of PAH degradation only, or to intermediates in the process also? Is this technically an observational portion to the study since the strain they were growing and analyzing was all under the same conditions?
What is the goal of this knockout? I feel like I am missing some information – is P73 a protein involved in RHD? So the knockout mutant would be unable to metabolize PAHs? It seems like this would be a lethal mutation, for a bacterium that can no longer utilize its sole source of carbon and energy.
So, in addition to the PAH degradation pathway, P73 can also metabolize many different sugars, but cannot initiate gluconeogenesis? I wonder what environmental factors would select for a bacteria with such a versatile ‘palate’ for carbon and energy sources?
A quick search revealed that Acetobacter aceti is a terrestrial organism, frequently sought out and used for its production of acetic acid from alcohol in fermentation of foods and wines. Makes me wonder how a gene that originated in this A. aceti ended up in sediment in the bottom of the Indian ocean!
This is the authors’ most interesting finding. A gene coding for a new protein that can initiate the ring hydroxylating dioxygenase step of the degradation pathway for toluene and biphenyl PAHs. As they state in the next paragraph, a marine organism that could degrade toluene would be a great tool for the cleanup of oil spills, since toluene is a toxic chemical that occurs naturally in crude oil.
BAD34447: large subunit of PAH-dioxygenase
CAG17576: ring-hydroxylating dioxygenase alpha subunit
ABW37061: Rieske domain; a cluster binding domain commonly found in Rieske non-heme iron oxygenase (RO) systems such as naphthalene and biphenyl dioxygenases
ABM11369: ring hydroxylating dioxygenase, alpha subunit
ABK27720: putative ring-hydroxylating dioxygenase large subunit
BAA21728: terminal dioxygenase component of carbazole 1,9a-dioxygenase
ABM11377: ring hydroxylating dioxygenase, alpha subunit
ABM11383: Rieske (2Fe-2S) domain protein
ABV68886: angular dioxygenase from fluorene-degrading Sphingomonas sp. strain
AF474963: Pseudomonas alcaligenes indole oxygenase-like gene, complete sequence; and putative dehydrogenase gene, partial cds
One question I could see the researchers pursuing in future studies is the control of the PAH degradation genes. Since P73 conceivably has the capacity to use multiple different carbon and energy sources, it would be logical it has some epigenetic capabilities to preferentially produce proteins to metabolize whatever substrate it was exposed to or growing on, right?
I understand that the mutants that didn’t produce a clear zone were chosen because it meant the mutagenesis process succeeded in mutating the gene. But why did they test it further with indigo if they already had what they wanted?
They concluded that the arhA gene was constitutively expressed because the the A4DR cells had low levels of arhA3 and arhA1 even when not induced. Could this be corrected by some of the methods we talked about in class like changing the inducer?
[These observations suggested that the genes involved in the initial oxygenation of acenaphthene did not function in strains AG2-45, AG2-48 and AG3-15.]
I get that a mutagenesis was performed and the strains were grown in different environments to determine whether they degrade acenaphthene. I don’t understand how the researchers cam to this conclusion.
I haven’t read about finding the regulator site in any of the papers we’ve read so far. How can knowing where the regulator binds be useful in further studies?
Could the OFRs that have no apparent connection to acenaphthene degradation have an unknown role in the cell? An experiment could be designed to knock out one to see if there is a change.
The scattered nature of the the sphingomonad’s genome seems to be an obstacle in identifying genes. What could be a possible explanation for its gene dispersal?
In the supplemental text, they found a lot of genes but I’m having a hard time relating them back to the initial goal of the experiment. Can you explain?
Oddly, these genes are in the same place. It hasn’t been this way with many of the other genes in the papers we read before. This is a guess, but I suppose different genus’s store their DNA differently than others.
In comparison to the other studies, it seems like the information found in this part didn’t go as far as the other papers. In other papers this sort of information would be used to physically test if the conclusion was true or not.
Is computational genetics a common way of discovering the function of genes? Did the information used in the other papers rely on this kind of work to point the researchers in the right direction?
This is a good point. The paper states that the pollutants are conversed to biomass, carbon dioxide and water. Which I guess either way you look at it, the remaining end products are less toxic or non toxic in comparison to the original pollutant.
Did they want to slow down the activity of their samples to make sure that the results they got were due strictly to experimental variables, and not the environmental changes while being transported?
Why did they choose to get sediment samples from 1-12 cm? Is this the depth where most of the oil had settled in the sediment? And why are the seawater samples taken from 15 cm? How big of a difference does it make in the results if these samples were chosen from different depths?
I was curious about this too. You would think they would be closer in range. Thank you for the clarification. It is true that we are all similar, but all so uniquely different.
I was interested in the GC-FID method, because I wasn’t sure what this was, so I researched it. I read that it was developed for analysis of oleic acid and fatty acids. What do these have in relation to nephthalene-degradation?
I am interested to in how the gram-negative can tolerate PAH better than gram-positive. I’m assuming it has to do with the peptidoglycan layers of the gram-negative.
Is the isolation of the naphthalene-degrading bacteria process similar to that which we performed in lab. Just test using different agars that they may possibly grow in until they get pure cultures?
Since this paper is saying that marine bacteria may be more significant in degrading PAH’s than terrestrial bacteria…. does this mean terrestrial degraders are limited only to land and vice versa, or are there some bacterial PAH degraders that are present in both marine and terrestrial environments?
The genomic library is used to represent all of the genomic DNA from an organism. This is done through DNA extraction in which the DNA is then cut to certain sizes. They are then inserted into vectors which are taken up by hosts. These hosts can then be used to amplify. The genomic library is used for sequencing genomes.
I looked up what agarose gel electrophoresis was, and I read that once a DNA molecule is digested, this method can then be used to see the fragments, because the electrical current moves the molecules across the agar.
Today in lab, when you drew the picture and explained how the clones can be an “overlap” of a region really helped clarify the idea of having the same 10.5-kb Sau3AI fragment.
I think the significance of not finding the gene clusters for PAH degradation would be that the organisms lacking the gene cluster could not degrade PAH’s.
What is the importance of the major cluster? I get that it shows divergence of a-subunit sequences and similarity of amino acid sequences, but what exactly is meant by the major cluster?
I was wondering the same thing, especially since they included “surprisingly”. I mean they did have aerobic As(III) oxidation activity, so why would there be no authentic PCR products?
Aren’t cyanobateria a type of photosynthetic bacteria? If that is the case are only photosynthetic bacteria the only type of bacteria of interest when it comes to arsenic use?
I researched this after reading your question. DNA libraries are made from fragments of DNA. Where cloned libraries are made from cloned, reverse transcriptase of mRNA. So cloned libraries do not actually consist of DNA sequences.
Going through this figure today, it really helped see what was actually going on in the figure. While the Arsenite is in light it is being oxidized, then when it is in the dark, it is being reduced.
Even though we went over this figure in class, I still don’t understand why they used the three different variables of 100% H2, 2mM sulfide, and no added electron donors. What are each of these three variables offering us? I understand that they are driving forces for reduction, so is it just offering us information on which is the greater of the three for Arsenate reduction?
I was wondering the same thing, because here it doesn’t really help us to get a clear conclusion on specific microbes. But I guess that could allow for a follow up experiment using the techniques we will discuss on Tuesday.
In response to Rhyann’s comment… Is it because photoheterotrophy was responsible for the oxidation, and acetate was the electron donor for photosynthesis?
In my opinion this is the most interesting paper we have done this semester. I enjoy reading about the links made between our anoxic Earth vs. oxic. And it now makes sense that more recent biological communities would share relation to As(III) oxidation and As(V) reduction.
So what is the point it “knocking out” the PAH degradation gene? It is just to make sure you are pin-pointing the correct genes involved in PAH degradation?
Is there normally a correlation between multiple antibiotic resistance regulators and PAH degradation pathways. I don’t remember reading this in our papers from last semester, but then I also wasn’t sure if I over read it.
In PAH degraders, is it more common for the PAH catabolic genes to be arranged in discrete or dispersed throughout the genome? I’ve never really thought about which arrangement is more common/likely.
Is the topic of interest here, why PAH dioxygenases can oxidize fluorine, but not use it as a sole carbon source? Or will we discover that along with determining locations of PAH degrading genes?
Thank you for sharing this Molly. Background information is always a plus. Dr. Ni Chadhain, what is GSL and LPS? I remember hearing you mention LPS in class today or Monday, but I can’t remember what you were referring to.
The southern blotting made much more sense when you went over it today in class. I did not quite understand about wanting two different sticky ends while also containing the RHD probe, but you clarified this concept.
So, from what you were saying in class today, this part of the experiment is basically just to tell us yes you can force the E. coli to make the protein, or no you cannot?
Are there different types of dioxygenase genes? Like does each PAH degrader have different types of dioxygenase genes or are all dioxygenase genes the same because they do the same job?
So are you saying that even though the RHD is present, it doesn’t mean that it will utilize the PAH as an energy source? Does this mean that the degrader will use whatever route is most catabolically favorable?
How do you choose which strain of E.coli you want to perform a transformation with? Is it just based on availability or are there certain strains that work better than others?
Homework 4 helped with understanding the information in Table 1. I understand what the table is actually talking about after performing our own BLASTs and listing our ORFs.
Can you better explain this sentence please? I’m not understanding how they can serve as substrates, but cannot grow on them. “Although strain A4 is not able to grow on naphthalene, phenanthrene, anthracene, and fluoranthene, these substrates could serve as substrates for the ring-hydroxylating dioxygenase system from this strain, since their corresponding cis-dihydrodiol products were detected in the resting cell reaction experiments.”
I’m surprised that monitoring PAH-dioxygenases in sphingomonads isn’t possible, because I feel like the species came up a lot in the BLASTs we did for homework 4, so it seems to be a well studied species.
I know that xenobiotic are something that is foreign to the body or environment. What is an example of a bodily xenobiotic? Are xenobiotics hard to degrade? I’m assuming degrading capabilities are based on what the foreign substance is.
When finding gene clusters of PAH degrading genes, how does this work if they are scattered out? Do they all have the same sequences, so when using certain primers, they all show up? Or does the DNA have to be cut multiple times in order to find every cluster throughout the genome?
I tried to look at the primers they used listed in Table 2, but it sent me to an online journal site, and I could not see the table because it only shows a preview of the article.
I think that transposon mutagenesis is really interesting, and it is probably my favorite part of what we have learned all semester. And I was really interested in our plasposon mutagenesis experiment in class, even though it did not work like we hoped.
Why do some strains produce little indigo formations such as strain A4-PCM1(pBBadA13), and other like A4-PCM1(pBBadA14) produce it significantly? What causes this difference? are they metabolically utilizing other things available in the agar first?
I’m glad that were are getting to perform RT-PCR in our own class experiments. I read on the ThermoFisher website that RT-PCR is so powerful that it can “enable quantitation of RNA from a single cell”.
How can they find the genes responsible for further degradation of acenaphthene if they are dispersed throughout the genome? How do you even determine which step is next in the pathway?
This could be a stupid question, but it has me wondering. Since this is concerning oil in the Persian Gulf, it makes me curious after the oil spill in the Gulf of Mexico. Could these Naphthalene-degrading bacteria be found in in the Gulf of Mexico?
Why did the researchers choose this particular oil spill? Is it a special type of crude oil, or did the environment in which it is found play a role in the selection?
Why did the researchers decide not to add an IR Spectrum to the analytical chemistry? It would help further narrow down any isomers that could be found in the tested substance.
Is the purpose of this research to identify PAH degraders or is the purpose to explore PAH degraders and their effectiveness in bioremediation of the surrounding environment?
Could PAH degraders be under-researched because of the lack of equipment or resources available for such exploration? 2000m is a long way down and we definitely cant send a human down there.
Later in the document E. Coli is used as a standard for unmarked DNA, does the first sample site act as the standard for the PAH degraders that are pursued in the second sample, or does this experiment assume PAH degraders are found dispersed throughout the sea floor?
I believe that a transcriptomic/proteomic approach would be useful, assuming that the researcher already knows the sequence that they will be looking for. On the off chance that their bacteria has a new sequence that has not been found, then the screen would surpass the gene and mis identify the bacterium.
With the fnr gene knocked out of E. Coli would this cause a completely anaerobic metabolism for E. Coli causing it to only use the Se as a source of carbon?
There is no further mention of the ogt gene in the rest of the results section so is it safe to assume that this gene played little to no role in the reduction of Se(VI)?
Based on this section E. Coli appears to naturally reduce Se(VI), but at a lower or less efficient rate than E. cloacae. Would this be the reason they did not try to incorporate the fnr gene from E. cloacae to E. Coli to prove that the fnr gene is responsible for the reduction of Se(VI)?
For E. cloacae to have a real world use in the reduction of Se(VI) that could reach toxic levels in an aquatic community, would there need to be a mutation of the fnr gene where Se(VI) is reduced in the presence and absence of O2 or would the rate of reduction be significant enough with out it?
Would these proteins be a way to make E. cloacae a suitable candidate for real world applications where a Se(VI) reduction capable bacteria would be needed for biosynthesis? Could these proteins be transplanted into the genome horizontally or would they have to be transplanted virally?
Wouldn’t this be a good opportunity for the authors to employ transcriptomics? What would be the benefits of employing the experimentation in the lab versus out in the field.
It is quite amazing how versatile rhodococci is and how it is able to degrade environmental pollutants that are too difficult to degrade for other organisms. While the rhodococci can metabolize xenobiotic, and halogenated compounds, which can be found in various herbicides. Is it possible for the rhodococci to metabolize other sorts of controlled substances such as pesticides or possibly fertilizers?
It is very interesting to learn that during degradation Rhodococcus uses aromatic rings that catalyze the initial reaction of bacterial degradation. During this degradation it uses two subunits, one of which is the alpha subunit that aids in electron transfer to the dioxygen molecule. But the other subunit, the small beta, was not explained further. Does this subunit have a purpose similar to the alpha unit? Or does it perform it’s own necessary task in the degradation process?
I think it is really interesting that the sample chosen was from an oil-contaminated sample in Japan. The fact that the oil could impact the growth of the culture compared to if the soil from that area was not contaminated. It makes me wonder what kind of oil was present in the soil and if it was contaminated with a different kind how would the affects differ.
I believe that OD600 is an abbreviation for absorbance or more specifically optical density. In this instance the sample was measured at a wavelength of 600 nm. This method helps in estimating the concentration of bacterial cells in the liquid.
I think it is really cool to be able to visualize the relation between the different mycobacterium and where Rhodococcus falls in the phylogenetic tree. It makes me wonder how other closely related would react under the same experiment or even their common ancestor.
I was thinking the same thing Aubrianna! It’s really cool to think that I am able to understand the paper better after every lecture and lab day. A lot of these terms and processes I had no idea what they were about but now I can formulate a general idea of what is happening during this experiment.
Since the Rhodococcus only exhibited an amplifies 100-bp band compared to its similar counterparts exhibiting a 78 bp band, would it not be expected that the Rhodococcus to perform the way it did? What is so different between these genes that allows Rhodococcus to do what it does?
It’s really amazing that a new naphthalene dioxygenase was reported in Rhodococcus during this experiment. Since it only shows a distant relation to the pseudomonas enzyme I hope to read more papers about this dioxygenase and its capabilities and be able to see what it is capable of in other situations.
It is really interesting that these PAH’s can be brought up the food chain and affect humans. I wonder what kind of effects they have on humans and what some of the symptoms would be to in order to diagnose a patient with being affected by PAH’s.
While it suggests that obligate marine bacteria may have more significant PaH degraders, could the physical parameters of their ecosystem be helping them? Terrestrial organisms have a restricted form of movement through their ecosystems where substances can only be moved via wind, water, or other organisms, but in marine ecosystems the movement of substances is much faster and less restricted possibly exposing the bacteria to more PAH’s.
I understand that the Marmur method allows DNA to be isolated from microorganisms and keeps the needed material active, and polymerized while keeping the majority of proteins and RNA away, but is there anything else about this method that makes it a necessity for this experiment?
So in this case, the important thing to remember is the fact that the converted products were being identified and measured as well as their purification that was produced in the culture supernatant, correct?
In the flanking region of phnA1b it is said that there is no present small-subunit gene in the oxygenase pair. Was this subunit lost in transformation? Also how does this hinder the genes functionality throughout the cell?
Tara, I also looked up P. stutzeri and found out about the spinal fluid and it’s similarity to the NADH ferredoxin. It’s really interesting that they are similar and it would be cool to be able to learn more about the history of the relationship between the two, just like you said.
I also have the same question Jesse. Is there anyway to access the Figures they are referring to in the text? I believe that if I was able of view them I might have a better understanding of what they are trying to say about their trees and how that relates to them falling out of the cluster.
Jennifer, I think it would make it more efficient. Since the PhnC is involved in both aerobic and anaerobic pathways, not only is it degrading the compounds fast but it is also able to degrade them when there is no oxygen is available.
Hey Alexandria, there are a bunch of other relationships between cyanobacteria and chemolithotrophs, and even some pretty large eukaryotes too. In some cases the cyanobacteria will provide its host with fixed nitrogen and the roles can even be switched around where the cyanobacteria is being provided for. So it sldo makes me think the same thing about whether or not they might be breaking down the arsenic to help each other out.
I was also thinking the same thing Louie. I can’t wait to continue reading this paper, it seems like a lot of the information will tie into some information from the Biology of Algae class that I took and how cyanobacteria could have nitrogen fixing activity during daylight hours if induced by low ammonium. I wonder if that sort of reaction will be shown in this different type of environment.
I agree Tim, identifying the protein produced would definitely help in figuring out which genes are responsible for the arsenic cycling or resistance quality since some of the PCR primers were not giving us enough information.
Caleb, I was also thinking about the environmental parameters of these anaerobics. Since the temperature range is so broad, it makes me wonder if they hold any sort of extremophile qualities, where their tolerance can withstand very cold or hot temperatures. It’ll be interesting to see the phylogeny tree and find out if they are related to any temperature tolerant organisms.
I wonder what kind of pigments these arsenic cyclers have, considering that they undergo anoxygenic photosynthesis it can be assumed that they use bacteriochlorophyll, but determining which kind might help with understanding the limits of their photosynthesis.
It would be interesting, considering the technology available now, if they redid the experiment with a omics approach to annotate the gene that oxidizes AS(III).
I was confused when I first read this paper as well. Thank you for your definition! This concept seems interesting, and I appreciated the explanations in the text to both co-metabolism and the use of consortium strains. These definitions helped me understand the biological aspects of bioremediation as it relates to microbiology and how it can be efficiently used to preserve the environment. It shows that there are natural ways that we can effectively target toxic oil spills instead of introducing harsh methods that could potentially devastate the environments even more.
I wondered the same thing, especially since there has been no further study for PAH degrading bacterial strains from the Nakheel region. Were there other factors contributing to the unique degradation patterns besides the low nutrients and high hydrocarbon pollution?
Why would conductivity of the sediment samples affect the PAH-degraders? Do conductivity and pH affect each other, or were they measured together for the sake of efficiency?
Does it matter how much PAH they end up with? It seems as if they would not have a large number of PAH left after this rigorous isolation process if they began with 1% PAH. It could be assumed that it would be a big inconvenience for them to repeat that process if they did not end up with a desired amount of PAH.
Why did they construct the phylogenic trees? Was this to organize and assess the complete variation of the bacteria present? Or did this help them in some way analyze which bacteria would degrade the toxic oils more efficiently?
What would the difference between the gram-negative and gram-positive bacteria have on the effectiveness of the PAH degradation? Why would that make a difference?
I was wondering the same thing. It would seem that LB would be more useful, but that conclusion cannot always be applied in such seemingly obvious situations.
Was this step in the experiment the reason why the phylogenic tree created earlier on in the methods portion of the paper? Does this make identifying effective bacteria and related bacteria easier? It seems that this could help find possibly both cheaper and mire effective cousin bacteria.
Since P. aeruginosa is possibly unsafe to test due to human pathogenic qualities, would there be any instance where that could be tested in a safe environment using technology? It begs the question of if it would be ethical or worth exploring other testing methods if it means finding the most effective PAH degrading solution, though it would be ethically irresponsible.
I had the same question, Cody. Does this mean that the other isolates can be defined as the bacteria that they are so similar to, or are the 1% and 2% differences significant enough for them to be labeled as something else?
It’s interesting to note the drastic difference of the 3 isolates on degrading pyrene vs phenanthrene. They all performed similarly on pyrene, but there are varying levels of effectiveness on the phenanthrene. In both cases, most of the isolates seem to follow the general pattern of the control and follow its fluctuations, just at a much higher degradation % rate.
That was also my first thought upon reading this section. After learning what turbidity and optical density are/consist of, it also made more sense that the solutions would be clearer if there were larger microbes present.
What is the purpose of the bacterial mat? It is mentioned here and in the next paragraph. Does it influence the temperature of the microbes? And would the absence of the microbial mats allow for more effective extreme cooling as apposed to them being present?
What is the purpose of using the different carbon isotopes 12, 13, and 14? I see that all 3 are used throughout the experiment. Do they relate to the bacteria being degraded, or do they relate to the methodology to extract the strains?
I agree, Kelsey. It seems as if the researchers obtained different results then they originally hypothesized. Once they identified the most efficient mineralization technique, they were able to continue experimentation to see which compounds would be degraded best by which means. It is clear in the graph in paragraph 2 that the 4567-24 has a much higher percentage of initial CO2 concentration captured.
It seems as if there are drastic differences in both the 14C-labeled PHE and the HPLC regarding the live cultures vs the acid-inhibited controls. Their effects are opposite of one another.
The PAH performed better with live cultures, but poorly in the control group.
The HPLC performed better in the control group and more poorly in the live cultures.
Was this effect to be expected since the PAH was removed in the HPLC?
What would the importance of determining the shape and size of the selenium particles formed be? Is this to determine how much of the selenium is degraded?
I had the same question. There didn’t seem to be a clear reason as to why one cosmic clone was chosen over the other or that any evidence of that would have been indicated later in the next paragraph or the rest of the paper.
Was the strain K-12 introduced to show that there are other means that E.coli can use to reduce selenate without the presence of the FNR gene?
Is this paragraph saying that the FNR gene was needed for the S17-1 strain and the YgfK,M,N gene was needed for the K-12 strain?
I would presume that the hydrocarbon degraders would degrade the oil differently, even if the oil in the surface waters is the same that is in the deep sea. The extreme environment of the deep sea would cause these bacteria to rely on other energy sources, possibly chemicals such as sulfur, etc. How much of a difference would this make on the effects of the hydrocarbon degraders?
I also made that observation, the extreme living conditions cause me to believe that these deep sea bacteria are a different strand than the coastal bacteria.
How would the phylogenetic identity of am organism determine its function, in this scenario with PAH degrading bacteria? I understand the concept of creating a phylogenetic tree for the purpose of identifying related and efficient PAH degrading bacteria, as conducted in the last paper. How would one determine the function of a certain phenotype or genotype of a PAH degrading bacteria?
How different are the microbial reduction of the selenate and selenite vs the abiotic reduction? Are they equally effective, or would one be much more effective than the other?
Does the process of Degradation depends upon Number and kind of micro organisms or the Chemical property of the PAH which needs to be degraded or both?
As Microbial degradation of PAH is quite interesting topic, I also read few other short articles about this, and I would like to share some information here. Prolonged exposure to chemical toxicants can cause adaptations in microbial populations that results in greater resistance to toxicity or enhanced ability to utilize toxicants as substrates for metabolism ( source of energy). Actual mechanisms for these adaptations are still unknown.
I wonder what if gene insertion(from other PAH degrading bacteria) is done, will be the affect of degradation of PAH increase or decrease or remains same!.
Why is there a differentiation in 48tRNA gene represent 19 amino acids and a tRNA for other amino acid? why not they considered 49tRNA and 20 amino acids?
I feel like the dark reductions occurred without a light source, which might cause it to have fluctuating temperatures as the experiment wore on from day to day. However, in the light controlled experiments, I think that the ability to control the temperature was easier as light was a component of the experiment!
I think the same! I wonder if the microbial populations were dependent vs. independent on light and if mutations had occurred in order to select for light oxidation and dark reduction.
It is definitely cool to see that hydrogen sulfide stimulated both the anoxygenic photosynthesis and in the dark incorporation of acetate. I think this makes sense because both reactions use sulfide as the beginning electron inorganic donor.
Since the bacterium rhodococci are capable of biodegradation of environmental pollutants, I wonder if it is capable of a mutualistic relationship with other organisms? Its ability to metabolize xenobiotic compounds that are not naturally occurring is quite unique and poses the question of whether it can exist symbiotically in different environments.
I was wondering the same? What is the specific function of the small beta unit? To my understanding, the alpha unit is involved in electron transfer at the iron catalytic domain. Does the beta unit function in reduction as well?
It is very interesting to note that the two ringed and three ringed PAHs are more readily degraded than the four or five ringed PAHs. I wonder if this has anything to do with the chemistry of the PAHs. Also, how can the enrichment be better modified to target four and five ringed PAHs?
I definitely noticed that too. I wonder if there was not continual enrichment and subculturing of the Rhodococcus on Fla would have resulted in different outcomes? What if the cells were instead subcultured with Nap or Phe?
It is very interesting that the attempt to enhance Nap, Phe, and Fla degradation actually resulted in their inhibition or had insignificant results. This can prove to be a beneficial discovery for producing a napthalene-inhibiting substance.
Since there was no homology between the tested rhodococci ARHDs in this study to any prior studies, does this quantify a new type of dioxygenases unique to this species of rhodococci? Are there any additional tests to confirm this?
I agree with this. It’s very encouraging to see education in real-world application. This studies shows that there are methods to inhibit degradation of certain compounds like Nap and methods to increase the biodegradation of Fla.
PAHs seem to be a by-product of factory and industry activities. Is there a way to regulate these processes or regulate the release of PAHs before it enters the natural environment?
Since Cycloclasticus operates in a marine environment, this bacterium must be salt-tolerant. Although I had no growth on the mannitol salt agar plate in lab 5, I wonder if I isolated an NP-degrader from a water sample if I would have growth on the mannitol agar and if it could possibly be a strain of Cycloclasticus?
I was wondering the same! Perhaps it may not have an effect at all? Maybe the presence or absence of this gene just serves as a comparison point for the two? I wish there was more information about the small subunit!
What does the data mean by “agreed well”? Does this conclude that, yes, the phnA1A2A3A4 genes actually did code for the holoenzyme of the aromatic dioxygenase? Is there an explanation for why the data may be a little off from the predicted sizes of Phn?
Since these proteins may be multipurpose electron transport proteins, would there be high regulation of these proteins since its functionality is enhanced?
I’m wondering if there is a specific reason as to why the activities of the fused, unfused, and sing-ring compounds were not different in these situations. Does it have to do with hydrogen bonding?
I’m wondering if these bacteria can use arsenate and arsenite as an electron donor in major pathways like glycolysis or the electron transport chain due to its high potential?
From the last sentence of this paragraph, are they suggesting that there are certain bacteria that can undergo “arsenification” and “dearsenification”? Also, what would be the ecological importance of forming arsenate and arsenite if they are toxic compounds?
Okay, so reading further, I do indeed see the the suggested coupling of arsenic metabolism. I wonder if one part of the couple is to recycle arsenic under anoxic conditions, will this also be tested under oxic conditions? Also, will the recycled products from this arsenic metabolism still have toxic effects on the environment or will they be neutralized?
I think that this is very cool that they took samples from rocks and cobbles that we would see on everyday hikes. This just reminds us of how much microbes rule the world! But on a more serious note, why was the slurry stored in the dark? Were the biofilms found in shady/dark areas?
I, too, agree that it is interesting that the sample was collected from a city in Japan. Why was Japan the site of sample collection? I wonder if there would be a difference if the sample was collected from an oil field in the Middle East. What type of soil and oil is optimal for the cultivation of PAH degraders?
I wonder why gas chromatograph was used as the method of analyzation. Do the bacterial strains give off a certain molecule as it degrades the aromatic hydrocarbons? Could a spectrophotometer also been used as a qualitative measure of analysis for bacterial growth?
In paragraph 9 of the introduction, it was mentioned that present study and isolation of Rhodococcus does not have homology with catalytic domain of any prior ARHD Rhodococcus species. In this study, it is reported that the isolated bacteria exhibited 99% homology with the Rhodococcus species in the 16S rRNA gene, which is a highly conserved gene region that is used to identify microbes in complex environments. I wonder, if this bacterial strain exhibited any homology with the Rieske center of ARHD’s as well. Does it also have homology with the catalytic domains of prior ARHD’s of Rhodococcus species?
It appears that Fla degradation followed a logistic rate according figure 2. I wonder how the degradation rates of Nap and Phe compares to Fla on a regression curve. I wonder what the difference in molecular structure led to the high rate of degradation of Fla in comparison to Nap and Phe. Knowing this, is there any way to select for the degradation of Nap and Phe better?
Is there a particular reason why the ferredoxin and ferredoxin reductase genes were amplified through PCR rather than another genetic set? Is this code unique to the bacteria?
I wonder how the results would turn out if PAH samples like from the previous paper and from our lab experiments were used instead. How would the collection of a variety of PAH samples have an effect on the results?
Is the degradation process of high-molecular-weight PAHs unclear because there is a lack of research in that aspect, or have researchers tried to study it but were unsuccessful?
If the P73T strain is the first fluoranthene-degrading bacterium to be found within the Rhodobacteraceae family, would that suggest that there may be other bacteria in the family that could degrade PAHs that we haven’t discovered or haven’t studied enough yet? Also, could the PAH-degrading genes be moved from P73T to other bacteria within the family using horizontal gene transfer?
What makes fluoranthene different from other high-molecular-weight PAHs that it is a good model? The previous paragraph mentioned that the degradation of high-molecular-weight PAHs wasn’t fully understood, so I’m curious as to why fluoranthene is the model of choice. I’m also intrigued as to why fewer fluoranthene degraders have been isolated from the marine environment. Is it because there is a lower fluoranthene concentration in those environments, or because bacteria marine environments aren’t the preferred research specimens?
I’m also wondering about this. Especially since the degradation of the high-molecular-weight PAHs is not fully understood. Some bacteria can degrade high-molecular-weight PAHs and some can, so is it the PAHs themselves that cause this, the bacteria doing the degrading, or both?
Me too. The fact that only a small percentage (like 10% I’m pretty sure) of the Earth’s oceans have been mapped and researched always has me curious about what else will be found in the ocean as time goes on. There’s so much to be found and researched from and about the oceans.
The mutant strain was generated to analyze and compare to the wild strain during experimentation. I’m curious to see if they will only try to examine the differences between the mutated strain and the wild strain, or if they will be trying to amplify the PAH-degrading genes in both the mutant and wild strains to see how the mutation will affect the function of the gene.
If fluoranthene is a PAH and is the only source of carbon and energy for the bacteria, then the mutated strain should starve and die since it is unable to degrade the fluoranthene. I’d like to see if this will be the case, or if the mutated strain will have an alternative PAH-degrading gene that will allow it to survive.
I thought it was really interesting that so many horizontally transferred genes were found. The P73T strain using HGT to adapt its PAH degradation is not something I expected, as I didn’t ever consider bacteria able to quickly adapt like that.
This answers a previous question of mine. I was curious to know if the bacteria with the mutated gene would be unable to use the fluoranthene, or if they would have an alternate PAH-degradation gene that would be used.
This is the same thing I was wondering! I looked up a few things, but there are mixed answers from what I have seen so far. I would like to know a for sure answer as well.
So if non-sporulating aerobic rhodococci are commonly found as biodegrading environmental than could they be used more prevalently in the world to break down manmade pollutants?
I find that it is interesting how different types of bacteria take different types of stains. It is important that we can use stains to distinguish different types of bacteria.
I wonder how much Fla would have been degraded if the samples had been allowed to incubate for longer amounts of time. For instance, how long would it have taken for Fla to completely degrade.
I wonder why the Rhodococcus sp. was able to degrade only that much of the ILCO in two weeks. How long would it take for the Rhodococcus sp. to degrade more of the ILCO.
I find it interesting how the writer chose to focus on how PAHs are present in the marine environment instead of the terrestrial environment. Perhaps this paper will focus more on how PAHs affect marine organisms.
I would be interested in learning more about how this genus of bacteria, Cycloclasticus, can degrade petroleum PAHs. That seems like it could have important real-world applications if used in the right way.
I wonder why the Luria-Bertani agar plates were chosen for the plasmids and to why there were added antibiotics. Also, why was that concentration of antibiotics chosen for the plasmids?
It is interesting that the researchers were able to show that PhnC is involved in the upper and lower pathways of degrading naphthalene, phenanthrene, and biphenyl. Since it’s involved in the upper and lower degrading pathways does it mean that these are more efficient in degrading these molecules?
It’s cool that the PhnC is able to use both the upper and lower pathways to degrade naphthalene, phenanthrene, and biphenyl. Since PhnC uses both pathways does that make it more efficient in degrading naphthalene?
I like that this paper is different from the other two papers that we have done. It is cool that there are bacteria that are able to use arsenic to survive when it is so toxic to a lot of other organisms. It is even more interesting that the bacteria are able to use the Arsenate and Arsenite to create energy for themselves.
I am also interested in learning why their PCR failed. Perhaps it was because the primers were not right for the genome. I wonder how it would be different if the researchers had taken samples from a different biofilm that had similar bacteria. I am interested in learning about how these different bacteria work to get their energy since it is an anaerobic system that is light-driven.
Maybe the slurry did not need to be intact because it was going to be used for growing the biofilm on media so they needed it in a liquid state. But that just a guess.
I think it was interesting that there were only two main types of bacteria that were found in these biofilms. It makes sense that several types of Archea were found in the biofilm since the biofilm was taken from a harsher environment.
I found it interesting how the As(V) and As (III) had nearly opposite reactions when introduced to the light and dark regimens. I don’t think I was expecting that to happen.
It is interesting that Rhodococcus has the potential for such varied uses. I would be interested in learning about what kinds of applications it has in environmental remediation.
I agree with you that Rhodococcus could have great benefits for the environment. It would be great for the many uses of Rhodococcus to be recognized and well developed in the coming years.
Why did the YE initiate the early degradation of Fla? Looking at the rest of the paragraph it certainly sped up the degradation of Fla during the incubation period significantly.
It is interesting how the increase of Fla concentrations could be inversely proportional to is degradation as the paragraph does not make it very clear. But reading the other comments does help to clear it up a bit with how Fla could be toxic in high doses.
I find it quite interesting that Rhodococcus what able to degrade aromatic fraction of Arabian light crude oil and the aromatics and alkanes of motor oil. But in the present study why was only the aliphatic fraction of ILCO able to be degraded by the Rhodococcus?
It is interesting how a new naphthalene dioxygenase was discovered during this experiment in the Rhodococcus sp. This is cool as it only showed a distant sequence relationship with the Pseudomonas enzyme.
I am curious to see how the Rhodococcus could be used in the future for biodegradation of different hydrocarbons. This is a very interesting product but I believe needs further research to help develop the methods to effectively use it as a biodegradation tool.
Are there be any benefit to not converting some of the molecules to their cis-dihydrodiol forms or to having the other molecules stay in their monohydroxylated forms? From the reading it says that the second set of molecules mentioned stayed in their monohydroxylated form due to structural instability in the cis-dihyfrodoils. But could there be any other reasons for keeping or changing the molecules one way or the other?
How many of these kinds of degraders where found to have the ability to oxidize indole to produce indigo? Do the ones in our experiments use that since the colonies will appear blue.
Why would they not show the data for the Southern Hybridization? I feel like it that confirm the loss of the region that you would want to show that data?
This is because of the size. Plasmids are only approximately 10kb, while cosmids are for approximately 40kb. Now they could have opted for a Phage instead possibly because the phages are used for the same size, but plasmids are going to be too small.
So since they are studying a new strain here, I am assuming that they will be incorporating the techniques in the previous papers here to identify the genes and pathways?
So GIs are the areas involved in horizontal gene transfer correct? Would they not run into problems with this later if they are only studying these areas?
Why would they not isolate and analyze the bacteria from that location? Was it because of the lack of enough nutrients and the excessive amount of pollution?
Are the nutrients’ analyses vital to the decision of the bioremediation microbes selected to attempt to naturally correct the issues? Is that how biologists know from the get go if bioremediation is an option?
It’s interesting that the rod bacteria are favored over coccoid bacteria. I researched on the internet and a website said that the large surface area’s don’t necessarily mean a large adhesive force.
It appears that they are saying that FNR regulation isn’t necessary if certain proteins are present. So yes, I think they are saying there are possibly multiple ways to reduce Se(VI).
I was wondering this same thing. If we are exposed to PAH’s through digestion do you think that the way certain foods are processed could increase the harmful effects? Also, regardless of whether the food we are digesting is processed or not, it could still have been contaminated from the environment. While it is important in protecting workers who could potentially be exposed to PAH’s via respiratory uptake, I feel it is just as important for people to be informed that these are in foods we are ingesting.
I also find it interesting that these can be identified using the same techniques that we are going to have the opportunity to use in the lab. If we were to do this testing in an area with greater exposure to PAH’s do you think it would correlate with the rates of cancer in that particular area? This is something that I was interested in reading a little further into. This lead me to asking myself if these areas that have greater exposure to PAH’s are well informed of this and are aware of the effects they can have on the human body.
Is there a chance that a person can be contaminated with PAHs such as ingestion without you realizing it? I’m wondering if the effects are immediate or are they something that can sometimes be over looked, such as shortness of breath and dry cough. It is hard for me to believe that I have never heard of PAHs yet they are potentially everywhere around us and even in what we eat or drink. I see that it says prolonged exposure causes more serious side effects but once you are exposed to these do they stay in our bodies or does our body eliminate them?
I was also thinking this same thing. In our lab I’m interested to see what the difference in growth is between our diluted cultures and our more concentrated cultures. Along with this I am wondering the difference in R2A plates and MSM plates. Will they differ in growth, color, etc ?
I was also thinking about the amount of PAHs in the environment. It it alarming that Dr. Ni Chadhain found this just simply in her neighbors yard. Is the public aware of things like this? I feel that there should be more awareness of the harmful effects PAH’s have and also more awareness of their abundance in the environment.
After reviewing the results I was wondering the same thing. I feel as though this is a question we will be able to answer after completing this course. After finding out what differs in their molecular structure I feel that we would be able to identify a better way to detect for the degradation and not only these but others as well.
It is interesting that I can immediately relate this back to what we are doing in our lab. Although, we are only a few weeks in to our course I already can identify what the difference is in growing a medium in an MSM plate vs an R2A plate. Even before we began this weeks lab we were told to look for a blue/black colony on our plates.
I did not initially expect the results with the added YE to completely inhibit the degradation of Nap. This makes me wonder if there are any studies about different strains that could possibly be tested on Nap. While this study was successful in regards to the degradation of Fla, I feel that there should be more experiments done to see what strains could be successful with degrading things like ILCO. Obviously there is going to be a lot more to the composition of something like ILCO, but I think it would be worth the extra work to help protect our environment.
I am curious what exactly (with the addition of YE) caused it to be so successful in degrading Fla, yet completely inhibiting the degradation of Nap. The Fla degrader seems to be more complex as they stated that another study found there to be two clusters depending on their PAH utilization capabilities. Do we know which cluster the one used in this experiment belonged to? Which also makes me wonder would a strain from the other cluster have been as successful in this experiment?
I guess I expected there to be a direct relationship between the concentration and the amount of degradation. Also, in this section of the paper it doesn’t state what the time frame was for the 750 and 1000 mg1-1 degradation but assuming that it would also be 8 days, would a longer time frame allow this to have a more successful rate? Or does it reach a type of thresh hold
I am interested to compare and contrast between these and the Rhodococcus that we read about in paper 1. The first question that comes to mind is wondering if one of these are generally more affective than the other? I am also interested to see the difference in the results of their degrading ability on different PAHs. If these are used in costal marine environments is there a chance that they have been used around our area for any type of bioremediation?
If marine bacteria is potentially more successful at PAH degradation this makes me think that Cycloclasticus will be the more successful one when comparing the degrades from this paper and the last paper. Also, it seems that the success of their degradation might be effected by what type of environment they are in, and not only which strain etc. Would we be able to conclude that one environment supports PAH degraders better than another?
This is the overall question I have had since the beginning of reading these papers. In just a short time we have learned the multiple routes and the extent to which PAHs can effect humans, animals and the environment… yet before this course I had never heard of “PAHs”. This raises the question of how many people are aware of the extent to which we are exposed to them and also the extent to which they effect everything in the environment surrounding us.
I was also wondering this. I think it is realistic to assume that depending on the situation one may be more effective than the other. But when they describe either one as effective do the byproducts play a role? Furthermore, does one give off more toxic by products than the other? I feel as though this is definitely something that they would consider when using either Fla or Cycloclasticus in bioremediation.
Will we get to learn more about the specific endonuclease that they are using to do this experiment? This also makes me wonder over what range can endonucleases be used or if they have to be very specific to the DNA sections that they are cleaving. Also, what was significant about the size range of the fragments that they recovered?
When I think of centrifuging I always think of chemistry 131 lab. I never expected further education to reveal the importance of this and that we can actually separate cells by simply centrifuging. After reviewing our pre-lab it’s so cool that we are going to get to do this Wednesday! The idea of us digesting the RNA and being left with DNA from our own culture is exciting and it’s also interesting to relate what we our doing in our lab directly to these papers.
I was also wondering the same thing after reading this paragraph! I do feel as though it is saying that there would be a specific DNA sequence that gives microorganisms the ability to degrade PAH, but I could be misinterpreting the wording. So does the order of the genes have any effect on their ability to degrade PAHs? Or does it matter that the order of the genes were different from the previously reported genes?
I was also thinking this! I’m wondering what the effects of the opposite direction of ORF7 is in the homology since later in the paper it states that the order of the genes (that exhibited high degrees of similarity with the polypeptide sequence) is different than the order from that previously reported.
Since they are located on separate transcriptional units does that completely determine that the electron transport proteins are shared with other redox systems or is it just assumed that they are shared as the reason for why they are on separate units?
When I was reading this paragraph I also was wondering about what degree of significance evolution would have on this expression mechanisms. Since it is so highly organized I feel that this is something that would be very interesting to read on from an evolutionary perspective. Could scientist alter the gene expressions to see what increases and also decreases the expression of the specific genes we see here?
I am interested to know if the conditions of Antarctica and its wildlife have changed in the past year, with less travel/tourism and possibly less exploitation of natural resources that leads to spills; and if any change has been for the better for the organisms that inhabit it.
Why and How is Diesel oil the most commonly used fuel in Antarctica? I have always known the continent to be a “cold and snowy desert” with nothing and no one around.
I wonder why they chose to only take 4 samples from each site (exposed and not exposed to diesel fuel). I would think they would want to collect more samples from the sites to have a larger sample size.
I find it interesting that S. xenophagum did not show a chemotactic response toward any stimuli used. Is this due to it not having the proper mechanisms? Does this microbe have chemoreceptors that can detect cues? Does it have flagellum that could help it move toward a cue? I tried to Google this strain to see what the cells looked like but was unlucky.
Wow! I think it is very cool that Salicylic Acid is produced in this process. This is a chemical that I am very familiar with in my day to day life (in skincare) and I know that many people are familiar with it’s close relative- Acetylsalicylic Acid aka Aspirin.
I think it is interesting that other strains have been isolated in other locations that have cold environments and it makes me wonder if there are strains that work similarly in hot to very hot environments.
How is Marine Oil Snow formed? Is it just an accumulation of oil particles that float back down from the surface of the spill? If so, is it possible to prevent this from forming by cleaning up the surface spill quicker?
Does the mineralization of hydrocarbon contaminants have any lasting effects on these environments? Is the product of this process not considered to be a contaminant or is it turned into a useful product for the environment?
I think it is referring to the controls that were killed with acid. But I do wonder why acid-killed controls were used rather than an E.coli strain as used in the last paper?
This was the comment I was looking for! The Meselson and Stahl experiments that we learned about in genetics were exactly what popped into my head when I read about heavy versus light DNA.
“Clearing”… is this similar to the Casein Hydrolysis test we did in Lab 5. Does this show the presence of caseinase exoenzyme production or something that has the ability to clear agar in the same way?
To add onto your question, I would also like to know if we have found this species or similar species in other ecosystems (like on land or in extreme conditions) and if they have the same roles in these areas or if maybe PAH degrading is something only the ocean-stricken species has adapted to?
How common is it for a microbial habitat to shift between an oxic and anoxic state in a 24 hour period? Is this similar to the day and night changes of plant photosynthesis and respiration?
I think it is interesting that this process has been found or studied in multiple very different sources: periphyton, which is located the vegetation found on the bottom of freshwater systems; the soil in Japan (assuming this is not from the sediment of a water system); and hot spring biofilms.
I think this idea comes from haloarchaea being around for a long time, and the process of anoxygenic photosynthesis comes from early earth being anoxic.
I would be interested in further understanding the process of endpoint determination using liquid scintillation and why this was used to measure mineralization.
As with the last paper and although I do not fully understand these processes, I enjoy reading about real-life experiments that correlate with what we are doing in our own lab – I recognize those bacterial primers!
Yes, I think it may be dominated by Ectothiorhodospira because of the dominance of it’s ability to cycle arsenic, it may overpower any other organisms ability to cycle the arsenic.
I am just wondering why these various and specific locations for the soil, water, or sediment samples. I understand that the samples that were collected have various levels of salinity, because the bacteria needs to thrive in hypersaline environments as said in the introduction, but is there another property that the samples need to have in order to be used for their experiment?
The Lowry method sounded familiar, but I could not recall what exactly it was. So I googled it and found that the Lowry protein assay is a biochemical assay for determining the total level of protein in a solution. The total protein concentration is exhibited by a color change of the sample solution in proportion to protein concentration, which can then be measured using colorimetric techniques. This now makes sense as to why they used this method while using the UV spectrophotometer.
The fact that these archaea have been found to degrade both aliphatic and aromatic hydrocarbons in high-salinity environments leaves me to wonder if they can also degrade beneficial compounds to these type of environments? Or is it just exclusively degrading the hydrocarbons? What would be the cons to exposing these fragile environments to more bacteria and archaea?
I find the topic of bioremediation very interesting. The use of naturally occurring or introduced microorganisms to consume or break down environmental pollutants. This should be the first step when cleaning up pollutants instead of introducing more harmful chemicals in order to clean up other harmful hydrocarbons. This is especially important for the Gulf of Mexico. The Gulf has a relatively high number of oil rigs, while also being the home of many estuaries. This can be a dangerous combination.
[Approximately 40 to 60% of the added [14C]benzene was converted to 14CO2 in a period of 3 weeks (data not shown), further suggesting utilization of benzene as the carbon source.]
This is good news in the sense that over half of the added benzene was converted to CO2 in a period of three weeks. Which is relatively fast working in the world of science. I just need clarification on this part, as the strain Seminole degrades benzene, it is stated that benzene is converted into CO2. Does this mean as Seminole uptakes benzene, it releases CO2? Correct me if I am wrong, but could that just add more problems to these estuarine environments?
It is said that the pob and pca genes are found clustered together in a contiguous pattern or scattered over several portions of the genome. I wonder is it better for these genes to be clustered or scattered? Or does it make no difference to the expression of these genes? I happened to google it, but could not find any reliable information to go on
I am curious as to why the organism was not able to grow on catechol if it was predicted to be an intermediate in the benzene and toluene degradation pathway. I understand they didn’t find a conclusion for this, but is there a specific test to find out why? Or could it possibly be a mistake on their part when making predictions about the organism’s genome?
I would be interested in finding more out about the genes that they predicted would show during the degradation process, but didn’t. I am assuming once all the genes are well known, that is when we can start to build a plan towards degrading the compounds that are harmful to marine environments.
I am interested in learning more about fluorene degradation. Being in marine sciences, the main focus is usually carbon dioxide, methane, nitrogen, and other major greenhouse gases. People rarely think about other compounds that come into play as well. Then to learn about microbes that can degrade these toxic compounds and the science behind it is super interesting,
I looked up the bacterial species as well and found that there are 8 halophilic and 10 halotolerant different strains for this particular bacteria. Im curious to see any similarities with the last degrading halotolerant bacteria we discussed for paper 1
I was wondering why they mentioned that they used phosphate buffer instead of Tris buffer. So I looked up the difference of the two that might lead as to why they choose one over another. Tris buffer is used as a running buffer in agarose gel electrophoresis to identify a species by looking at the base pairs. On the other hand, phosphate buffer is used in isolating cells from tissue to maintain pH and keep cells alive. I also saw where phosphate is more expensive, but I see why it was needed instead of the Tris buffer for this experiment.
I like how detailed these authors are with their experiment. It makes it easier to read and understand when they explain each step and what the product resulted after doing each step and not just assuming the reader knows how they got to a specific result.
Coming from a now second time reader of scientific papers.
I think it is interesting how we are now on our third paper about degradation of contaminants in the marine environment by naturally occurring or introduced organisms, and I have never heard anything about it beforehand. Introducing these organisms to polluted areas seems like such a good idea. I wonder if this is actually taking affect anywhere around the world
I didn’t know about the cre-lox recombination method so I did a little research. Cre-lox recombination involves the targeting of a specific sequence of DNA and splicing it with the help of an enzyme called cre recombinase. Cre-lox recombination is commonly used t circumvent embryonic lethality caused by systematic inactivation of many genes.
I think it is interesting that they took samples from the Indian Ocean sediment, but then grew the strain on artificial sea water. If this is to work in the marine environment, wouldn’t it be more beneficial to take ocean water samples as well instead of something man made?
Could you further explain this figure? There is so much small information, I am not sure what to take out of the circular map. I am also confused to what the inner circle is showing
This is the most genes involved in the degradation of PAHs in a single strain that we have read about. Does this mean that the P73 is more capable of degradation of hydrocarbons than any of the other strains that we have learned about in the previous papers?
The variety and number of tests performed should show relationships among naphthalene degrading bacteria. Are there any other tests that would help with the study? Is there any particular test that is more beneficial or conclusive in determining naphthalene degrading bacteria?
Since strains N2, N7, N9, and N10 are all phylogenetically related (from figure 2), one would assume their growth rates would be nearly the same; however, the growth rates range from 0.345 to 0.928 (from table 1). Why is their so much variation?
Why is it that 400 ppm was the optimum concentration of naphthalene? Is it more important simply to understand that 400 ppm is the optimum concentration of naphthalene?
I find it interesting that in paragraph 4 gram positive bacteria were isolated and found to play a role in naphthalene degradation, but in this paragraph, gram negative bacteria were favored for naphthalene degradation. Does the difference have to do with the lipopolysaccharides in the gram negative bacteria’s outer membrane?
Since bacteria were isolated from an oil polluted area in the Persian Gulf, could bacteria be isolated from the Gulf of Mexico which was polluted during the 2010 BP oil spill? I think it would be interesting to isolate bacteria from a waterway that most people in this region have encountered.
I know some of the main points of the study were to isolate and classify naphthalene degrading bacteria, but unless I missed it, the paper didn’t discuss how to insert these degrading bacteria into oil polluted environments for controlled bioremediation processes. Is this being tested?
Various chromatography techniques were performed in the method of this lab. Chromatography is useful in separation by size, charge, and affinity each of which is useful in identifying a pure isolate.
Various chromatography techniques were performed in the method of this lab. Chromatography is useful in separation by size, charge, and affinity each of which is useful in characterizing a pure isolate.
Since naphthalene is a two ringed structure and phenanthrene is a three ringed structure, is it important to know which bacteria can use which chemical? Or is the fact that they have the ability to degrade either and/or both of the compounds the topic of interest?
The Sau3AI fragment of DNA is the gene which allows pH1a and pH1b to oxidize certain aromatic compounds. Is this inerpretation of the paragraph correct?
What is the significance of not finding the gene clusters for PAH degradation on the plasmid, and why doesn’t the strain have a plasmid at all? This is a unique characteristic as plasmids often contain genes for antibiotic resistance, degradation properties, etc.
Monooxygenases are enzymes that transport one oxygen atom from the air to the substrate, and dioxygenases are enzymes that transport both oxygen atoms from the air to the substrate. I think this comparison is useful if, like me, you have trouble keeping these definitions straight.
If other dioxygenase genes in Cycloclasticus were examined, what would those results conclude? Since this paper is several years old, I assume additional research has been done.
How are some organisms able to utilize arsenic, but for others arsenic is toxic? I understand that if an organism can utilize arsenic it does so by using it as an electron donor or acceptor, but what determines whether or not an organism can use arsenic?
Alkalithermophiles are extremophiles that are grow optimally at high pH (around pH of 10). The rod-shaped bacteria, Anaerobranca californiensis, is an example of an alkalithermophile. From further research, I found that this bacteria grows best in the presence of elemental sulfur, polysulfide, or thiosulfate which are found in the spring waters.
A radioassay tests a radioactive sample to determine the intensity of its radiation. Incubation is a process that maintains a favorable temperature and other conditions promoting development. In this experiment, is it correct to deduce that radioassay incubation was used to “grow” the bacteria?
Assimilative reduction is where an element is consumed and incorporated into new cell material. In light-incubated slurries, assimilation was not stimulated by the presence of arsenic oxyanions. Why is that?
Senescence or biological aging is the gradual deterioration of function characteristic of most complex lifeforms. Essentially the third sentence is saying growth rates decrease because the microbes are aging and toxins are accumulating.
Sulfide and hydrogen increase the rates of arsenate reduction because they are at the top of the standard reduction potential chart. This means they give off a lot of energy.
Fluoranthene is a fusion of naphthalene and benzene by a 5 member ring. An alternate PAH is one where the pi centers are not adjacent to another pi center. In a non-alternate PAH, somewhere on the ring there are pi centers that are adjacent to one another.
From further reading of the paper we will find the answer to the following question, but for now, could it be guessed that the fluoranthene degradation pathway of Celeribacter indicus is initiated by dioxygenation as said in paragraph 2?
One of the goals of this experiment was to isolate the genes needed for PAH degradation. Should genome sequencing help to determine the fluoranthene degradation pathway?
I had never heard of the term silylation, so after a quick google search, I found that this is replacing a proton with a trialkylsilyl group. The purpose of this is for analysis using GC and mass spec. How is this helpful in our understanding of the fluoranthene degradation pathway?
I’m having a difficult time understanding why strain B30 and strain P73T were compared. Are the two strains more or less the same other than B30 lacks most of the RHD genes found in P73T?
Are there other genes throughout the genome that could also be responsible for the integration of foreign sequences needed for HGT? In this paragraph it talks about region B, but what about regions A, C, and D?
Knocking out a gene is a good way to determine if a gene has a suspected function. In this approach, gene function is studied by examining gene loss. What other methods can one use to determine a gene’s function? Genomic DNA analysis?
Why is aromatic ring hydroxylation the most difficult step in fluoranthene degradation? Is it a difficult reaction to get started, does it make unnecessary side products/reactions, or is it something completely different?
Horizontal gene transfer is a good way for new genes with new functions to be inserted into a genome, but can HGT be harmful to an organism? Can it insert a gene that inhibits a necessary gene? I assume such organisms wouldn’t survive in nature since this would decrease the organisms’ fitness.
Sphingomonas sp. strain LB126 is unique from other microbes we’ve looked at since it was isolated from PAH-contaminated soil. We usually look at microbes from PAH-contaminated marine environments. Is there any difference among microbes found in soil vs in a marine environment?
I did some googling on degenerate primers. I found that they’re useful when isolating the same gene from several organisms, as the genes are most likely similar but not identical to one another. Degenerate primers are also used when primer design is based on protein sequence.
From what I found, sonication involves applying sound energy to agitate particles in a sample. Some of the purposes include: to break intermolecular interactions, for the production of nanoparticles, to disrupt or deactivate a biological material, to evenly disperse nanoparticles in liquids, initiate crystillization, loosening particles adhering to surfaces, etc.
For this experiment, is sonication used to break up the pellet into the liquid?
I’m trying to make sense of the information found in paragraphs 6-8 and in Table 2. From what I understand, this is to compare what substrates FlnA1-FlnA2 can use to which substrates two common dioxygenases, DFDO and CARDO can use. Are FlnA1-FlnA2 genes that were found in Sphingomonas’ genome that function in fluorene degradation? From these 3 paragraphs and table 2, what exactly should we find to be noteworthy?
Cells use glucose in glycolysis to produce pyruvate. Pyruvate can then be broken down to lactate in lactic acid fermentation, ethanol in ethanol fermentation, or acetyl CoA in the citric acid cycle.
Shouldn’t we expect that fluorene is not the best carbon source since smaller PAHs such as naphthalene are easier to degrade? Or am I confusing this with the fact that smaller PAHs are more widely studied as they’re model PAHs?
I looked up the 16 priority PAHs. From what I found, these PAHs are of high priority because of their potential toxicity in humans & other organisms and their prevalence & persistence in the environment.
From what I found on google glass power is used for purifying fragments from agarose gel electrophoresis, is that correct? What is the purpose of using glass powder?
Shotgun cloning is different than shotgun sequencing. Shotgun cloning is used to duplicate gDNA by fragmenting DNA with a restriction enzyme. These fragments are then cloned into a vector. In shotgun sequencing, DNA is randomly fragmented with a restriction enzyme, but then, the fragments are sequenced. These sequences are reassembled on the basis of their overlapping regions.
In this paragraph, they used targeted mutagenesis which relies on homologous recombination. I’m going to attempt to explain it, so please let me know where I explain it incorrectly. In this type of mutagenesis, the arhA1 gene was cloned into a suicide vector. The antibiotic resistance gene (Gm) is inserted into the arhA1 gene (in the vector) which disrupts its function of PAH degradation. Homologous recombination occurs, so now the mutated arhA1 gene is inserted. This allows the gene to show antibiotic resistance, but not PAH degradation. You know you’ve successfully mutated the gene when the original function is inhibited.
So in this experiment, genes encoding for ferredoxin & ferredoxin reductase were on one of the plasmids that was transformed in E. coli. I know ferredoxin works in electron transfer, but what exactly is happening with it in this experiment? What is the purpose of having a plasmid with genes encoding for ferredoxin and ferredoxin reductase?
Transposon mutagenesis allows genes to be moved to the chromosome of your organism of interest. This causes mutation because the gene is inserted in the middle of a functioning gene. In this experiment, they’ll mutate the genes involved with acenaphthene degradation.
In paper 3, the authors found the genes involved in the initial steps of acenaphthene degradation, oxygenation. In this paper, the authors hope to find genes involved in the next steps of acenaphthene degradation and the genes involved with acenaphthene degradation regulation. Would sequencing the organism’s genome give you this information?
I tried to do some googling on what a xenobiotic is. From what I found, they’re substances which are foreign to the organism. Xenobiotics include drugs, industrial chemicals, naturally occuring poison, and environmental pollutants. This website might help to answer some of your questions.
I know that sometimes the order of events in the papers is not always written in the way they were carried out. Is that the case here after the DNA manipulations paragraph?
I’m assuming arhR is a gene involved in acenaphthene degradation. Disrupting the gene will give us insight into the function of the arhR gene. From this paragraph, are we able to determine what disrupting the gene caused to happen, or would this information be found in the results section?
I was unfamiliar with what a primer extension analysis was. From my google search I found the following:
Primer extension allows the 5′ ends of RNA to be mapped. It can be used to determine the start site of transcription by using a primer which is radiolabeled.
I’m not sure how old this paper is, but has further investigation of the degradative genes and regulatory mechanisms been done to help clarify the acenaphthene degradation pathway?
After the process of bioremediation, what type of effect does the end product of microbial biomass have on the environment? Does it show an effect at all?
What are the costs and effects of using different strains of bacteria for biodegradation? What difference would you expect to see when deciding between bacteria based on their biodegradation ability?
Looking at the data table, I am unsure of exactly what the numbers corresponding to each carbon substrate represent? I understand the table shows the utilization of each carbon substrate but I am unsure of how the numbers are calculated.
How would evolutionary pressures cause species of Pseudomonas putida to be scattered to different clusters and those from Sphingomonas to be in the same group?
Would there be a method to more directly determine which biofilm microbes were associated with these arrA amplicons involved with As(V) reduction in dark incubations?
My guess to your question based off the section is that V. fischeri ES114 contained something that the others did not which inhibited the growth specifically to that culture, because it was stated that the other culture fluids from the other tested strains did not inhibit V. harveyi growth.
I also noticed how well of a specimen vibrio seemed for the experiment, knowing this information makes me think the results of this study will be great in terms how much data they will be able to collect.
Reading the paragraph right above this one, siderophores have a high affinity for Ferric or Fe^3+ iron which is the abundant form of iron in at neural pH in the presence of oxygen, so I came to the conclusion that if it is not that specific affinity for iron then it would not actually bind. In conclusion not all siderophore producing microbes would not have the same affinity to iron.
I actually knew that nitrification played a role in marine life, but I was not aware of the significant role that it played until I just read the data. So, I also agree that this was a pretty interesting finding.
After reading your comment, I was curious and decided to research your question. From the looks of it, as if right now, they are not able to give too much more information other than that diversity is negatively impacted. So, I am guessing some life exists but not a lot.
Based on your definition and the paper’s definition of what a steady flux is, it seems to a constant considering it is distinguished by the amount of atoms crossing.
I think they were determined or at least thought to have made an appearance in the step above where it says “Anammox rates were measured by the means of isotope-pairing” and were then verified using the CARD-FISH.
“Hydrocarbon contamination in Antarctica has profound effects that have been shown to reshape the structure of microbial communities as well as affecting the abundance of small invertebrate organisms”. How exactly are the microbial communities reshaped? What is to be expected of the communities when they are encountered? Also, does that mean there is a reduced amount of small invertebrate organisms or more of a diversity when taking into account their amount?
In response to your question regarding limiting the amount of contamination, we would have to figure out what might be one of the biggest contributions in causing the contamination in order to reduce it. Because the causes of oil contamination could easily very. If that was done, then I feel confident that we could play a role in the reduction of contamination.
Finding your question interesting as to why biofilm may not occur in this experiment, I researched it. Based on my research, the formation of biofilm has to do with antibiotics, biocides, and ion coating. Those can interfere with the attachment stage in biofilm which will cause the other stages not to necessarily be applied.
I also wondered what the change in temperature would do to the buffer. Personally, I think it would make the 90 minute incubation drop, but what will the changes in temperature do to the results of the experiment? what direction will it go?
[ To this end, we assessed the ability of culture supernatants to emulsify diesel (E24 index) as well as the ability of supernatants to generate water droplet collapse by altering surface tension (Figure 3G). In both tests, D43FB supernatants lacked emulsifying abilities, and exhibited similar behaviors to negative controls, suggesting D43FB does not secrete biosurfactants to aid in its degradation of phenanthrene.]
While reading, I was not necessarily sure what “emulsify” or “emulsifying” meant. I researched the terms and they basically mean to have two liquids in a colloid and not completely mixed together. So, in other words, it now makes sense as to why emulsification had nothing to do with the degradation of phenanthrene due to the lack of emulsifying abilities.
I agree with you as far as not truly believing the D43FB will thrive when put into the conditions of Antartica. It seems as if they are more so of hoping for the results will turn out good solely because it had the best results when it came to the others, but the analyzation doesn’t seem too solid.
In addition to what you’re saying Ryan, I would also love to compare how S. xenophagum D43FB would respond in the situation in which the temperatures change drastically either much colder or warmer. Does the weather play a major role in how the isolate degrades and replicate?
I was not aware until reading the article that it was forbidden to bring foreign organisms into Antartica, so I was caught by surprise as well. Also, I feel like this should be enforced in a lot more other places as well because according to the data, strict laws such as this are what is making this environment such a great place. There are so many places that could benefit from the narrowing down of decontamination techniques/strategies.
My initial thoughts after reading was what necessarily makes iron a great source of competition as opposed to any other source and what lead them into making the decision as to why they should use it?
In response to your question, I do not necessarily think it shows evidence of that because of it stating the V. fischeri ES114 does not produce aerobactin and/or sufficient iron. In other words, it doesn’t clearly state if low iron is the sole reason.
I think it’s very interesting to see the drastic change when the oxygen has nicely depicted lines decreasing into the suboxic area then travels backwards increasing back into the lower oxic area. Then the H2S starts off at a plateu or a completely flat rate then gradually increases as O2 completely runs out.
I am a little confused in Figure 2D because what I am interpreting based on data is that the NOx- decreases in some areas of the lower oxic area while it slightly increases then reaches a rate where it plateaus and does nothing at all like it was at the beginning where there was no oxygen? So, is this saying that the oxygen ran out?
I definitely agree with your comment, without the genes being transferred in that manner, the experiment would not be as effective. I wasn’t aware of the horizontal transfer being across multiple lineages either until reading this section of the article.
Yes, this last sentence gave a great/more clear summary of what was actually concluded at the end of the experiment. Before reading it, I did have some confusion of what they were suggesting. I found this experiment interesting because I have never seen this type of interaction “occur in nature”.
I honestly don’t think it would be considering the genes were rarely seen to begin with, so I think it just happened to be abundant in that particular location based on the conditions. The only way I feel like would be observed is if they had similar conditions.
I would actually like to know the answer to that question as well, because I thought they initially chose the Black Sea due to its great source of microaerobic nitrification. If I had to guess though I would say that it would not be as directly coupled which it what would make the Black Sea so unique to begin with.
I found bioaugmentation very interesting. I found a study where they actually used bioaugmentation and biostimulation hand in hand and discovered that together these two processes were optimal for oil degrading. Interestingly, there are Antarctic bacteria that are capable of hydrocarbon degradation at extremely low temperatures and they concluded that the use of bioaugmentation in those situations could enhance the rate of bioremediation.
I found myself pondering similar things during this reading. My initial thought was that it would be doing more harm than good as far as bringing those pollutants over as well as any non-native species of microbes and things that are on or in the icebergs. I typically tend to believe that disrupting the natural order of things means it shouldn’t happen. However, with what I’ve learned about serious drought issues that people are facing I feel this is a fairly logical route to supply the people. The captain who conceived the plan seems to have an ecological, well- thought out plan to make it work. I think the pollutants would be in such minimal amounts that purifying that would become the easiest part of the process.
I don’t necessarily believe the experiment would have been hindered if another media was chosen that suppressed certain bacterial growth as long as there were runs made with a control media. However, this is obviously very dependent on which media is used.
When pondering which media (if any) I though would have been more successful, I found myself thinking that R2A is typically used with slower growing bacteria and can be often suppressed by faster growing colonies. This made me wonder if they came across this issue in the field. Unfortunately, I never really found the answer for which agar would be better than R2A. I would lve to hear others thoughts.
After reading about biofilm production as well as this paragraph, I was curious as to which of the 350 colonies produced were the ones producing the biofilm and if there is a method to differentiate between those working in biofilm production and the colonies who didn’t. If it wasn’t all of them.
I believe the answer is yes! When undergoing transformation, the cell lyses and releases those intracellular components into the environment. That includes DNA fragments which a bacterium could take up to gain resistance.
I was also confused. From what I understood when looking it up, I found that while most siderophores are strong enough to pull iron from host-binding proteins so maybe since the microorganisms producing them are in iron deficient areas its just another case of survival of the fittest we observe in countless environments. There are siderophores who outcompete others based on better genetics.
From what I looked into I understand it to be a more specific type of PCR (DOP-PCR). It allows the employment of olignucleotides of partially degenerate sequences specific for genome mapping. Olignucleotides primary use seem to be primers for PCR so your assumption is correct. As for specific reasoning, I found they have the ability to amplify a small amount of DNA as well as being designed for length and G/C content. They flank the target reason so it seems just to be a wonderful primer for this use.
I read that this type of assay seems to be the most accurate as compared to counting-based methods for plotting growth curves. Is this because the fluorescence- based method isn’t dependent on determination of total cell numbers, but more so assessing fluorescence of the given sample of single cell from the population at a given time? I feel that contributes to lower variation rates among the plated samples which in turn only increases reliability especially when assessing the lag, log, and stationary phases.
I understood it to be both. They kind of go hand in hand as you can predict antibiotics resistance based off of mutations. Mutant construction of essential genes aid in resistance. Or am I missing some context?
Specifically CAS because of its ability to detect various types of siderophores. I found a paper that was saying in comparison of solid versus liquid that the liquid reaction-rate was an positive over the solid in all species tested. So maybe it was easier to quantify this way?
[The inability to grow was not specific to V. harveyi: V. fischeri ES114 culture fluids also prevented growth of Photobacterium angustum S14 and Vibrio cholerae C6706, while no diminishment of growth yield of Vibrio parahaemolyticus BB22OP and Vibrio vulnificus ATCC 29306 occurred (Fig. 1B). ]
comparing this to figure 1B made for much easier understanding. However, I found it a bit confusing that the growth inhibition was specific to V fischeri ES114 while others still had the inability to grow. If someone could offer some clarification.
[In communities, public goods can be exploited by cheaters who acquire advantages through use of the good but who do not pay the energetic cost of goods production. In the context of siderophores, a cheater need only possess genes required for recognition and uptake of the siderophore-iron (Fe3+) complex. ]
This comparison helped my understanding greatly. I found it very interesting that the cluster of siderophore biosynthetic genes encode for the ancillary functions of reception and transport for the V. fischeri.
I know that this just saying that V. fischeri ES114 outcompetes the others by uptaking all the iron which in turn also deprives the other of iron. Would the cheaters not steal the siderophores to outcompete V.fisceri ES114 since it wouldn’t be paying the metabolic cost to produce/secrete?
[ This co-culture system, in which either or both species can be genetically manipulated, provides a route to the quantitative investigation of both competitive and cooperative interspecies interactions that occur in nature.]
The identification of aerobactin acts as an inhibitor or the V. harveyi while working towards eliminating production in the V. fischeri ES114 thus creating this co-culture. I found this so interesting and somehow it helped the whole discussion click easier for me. It’s cool that this experiment led to quantitative data for the competitive and cooperative interspecies interactions.
[ nonthermophilic MGI Crenarchaeota constitute a significant portion of oceanic picoplankton (up to 30%) (21, 22) and a considerable fraction are likely autotrophic (23, 24), it is speculated that these MGI Crenarchaeota could be more important nitrifiers in the oceans than the usually less abundant AOB (18, 19). ]
As a student who has been studying marine science, I found this particularly interesting specifically for its higher abundance. Nitrification in oceans is important to the nitrogen cycle. For several reasons, including the microorganisms that use it for nutrients. I am eager to read where this study went and infer the impact it will have on things I’ve learned this far.
I think nitrate levels increase with depth because there’s no producers consuming it as well as regeneration due to decomposition of organics and the hydrothermal vent systems.
I looked up what a steady state flux is and from what I read its a diffusion process distinguished by amount of atoms crossing a unit of area perpendicular to a given direction per unit of time. So basically it is a constant concentration gradient?
I noticed they got their water samples and examined physical properties. Chemical analyses, dark carbon fixation, and Anammox rates were measured. Presumably, this data could be used to provide evidence of the amoA activities in the water column from which the samples were taken. This also provides comparative data for the coupling between the nitrification process and anammox along side the chemical profiling.
In Fig. 1, I would say these data points would be expected. In part a it shows nitrogen levels tend to be higher/largely distributed in greater depths due to lack of light and producers as well as regeneration through decomposition of organics by bacterial colonies.
Part b: Oxygen is most abundant in surface waters and decreases with depth. Sulfide would increase with depth as we get deposits of decomposing matter settling down.
Part c: light transmission is obviously one that is higher in surface waters and dissipates with depth as the light is scattered or absorbed by particles floating in the water. The particles increase with depth and in return decrease light penetration.
Part d: Anammox bacterial cells are more abundant with depth as they love abundant ammonium and nitrite as well as dark, anaerobic conditions (which matches the graphs before). The NH4 spikes where there arent more dense colonies and decreases when bacterial densities increase.
Fig. 4 shows that the most efficient production of 14N15N and 15N15N are converted from 15NH4 (a) where both numbers steadily increase.
(b) From 15NO2, we get equal 14N15N, however, 15N15N was not produced.
(c)Finally, with 15NH4 + 14NO2, we see 15NO2- produced to equal amounts, but drop off. And 14N15N had steady production, but the numbers weren’t quite there. Also showing no production of 15N15N.
[In accord with previous findings, dissolved oxygen in the central Black Sea (43°14.9′N, 34°00.0′E) [supporting information (SI) Fig. 5] decreased from fully oxic to <5 μM at 85 m (σt = 15.83, for comparison with studies in other parts of the basin) (Fig. 1). The suboxic zone extended from this depth to 112 m (σt = 16.15) below which sulfide started to accumulate. ]
I was expecting a similar discussion to this based off of just the graphs, however, I was not as well spoken. Although I falsely assumed the graph down to an anoxic level instead of just a suboxic zone. The sulfide was predicted to increase in the suboxic zone because of the sulfur-reducing bacteria and their tendency to thrive in a lower oxygen environment.
[ The total NOx − production in the oxic zone (55.5 μmol of N m−2 day−1) also could not match this NH4 + loss because of anammox, which consumes 1 mol of NO2 − per mol of NH4 + oxidized. Therefore, an additional local source of NO2 − and/or an additional loss of NH4 + must be present to reconcile the difference. ¶ 16 Leave a comment on paragraph 16 0 The best candidate to explain this phenomenon is microaerobic or anaerobic nitrification, whose direct coupling with anammox, the so-called completely autotrophic nitrogen removal over nitrite (CANON), has been demonstrated in bioreactors (35, 36). At 100 m, nitrification was evidenced by the production of 15NO2 − (12.9 nM day−1) in incubations with 15NH4 + + 14NO2 − and no measurable oxygen (Fig. 4).]
The coupling of anammox and microaerobic or anaerobic nitrification was apparent when analyzing anammox rate measurements and the isotope pairings in Fig. 4. I just found this part of the experiment particularly interesting because it gave me more insight into whether NO2- in marine water columns comes from the nitrification process, nitrate reduction, or both. Directly ties into my studies in physical oceanography.
Good question Justin, I was thinking the same thing. I think that it does generally occur when smoke enters the air, rather it be burning firewood or smoking a cigarette. PAHs are found in a various amount of things that we tend to burn, therefore I feel a common way they get exposed to our environment is through smoke in the air.
I agree with you Dawson! I find it intriguing myself how much science and new technology have advanced allowing us to isolate and identify one bacterium from another. I find it interesting that we can isolate these enzymes and further manipulate them to see what they can do.
Why is it that more information is available on bacterial biodegradation of lower molecular weigh than of high molecular weight? What factors play a role in performing microbial degradation? Are these processes also harmful to the environment?
Is the Persian Gulf still suffering from being polluted in 1990? How does the PAH bioremediation process converge the pollutant material into a useable substance that is not harmful to the environment? Can the marine environment actually benefit from these strategies?
Why did the samples have to be incubated for a seven day period? How could they actually determine the amount of growth if the samples weren’t being directly measured?
What does the action ” agitating in the vortex” mean? How does this process of using gas chromatography work? How long does it take? Is this the most accurate way to calculate the culture medium?
What caused the strains show a dramatic decrease in growth after 400 ppm concentration? Why did the Salegentibacter sp. strain N7 have the maximum growth.
What is the process on how the activity and adhesion were examined? What was different about strain N1 that allowed it to have the highest values of emulsification activity and values of cell surface hydrophobicity?
What does the 16S rDNA sequence control? Why was it necessary for them to clone the catechol meta-pathway operon genes? What characterizations did the strain contain that was necessary for the conclusion that it is a good model organism?
What other organisms belong to the genus Sphingomonas? Did all of the results show that the strain should be apart of this genus or were there specific traits that made them think this?
Is there any relationship with the distance of spacing between encoding genes? When it says the arrangements are similar to others, does it mean they function the same?
What is the ideal or preferred structure of the meta-pathway operon? How do the differences in structure alternate the understanding of metabolic capacities?
Why weren’t they able to obtain authentic PCR products for the arsenite oxidase genes? Was there an error? Would they normally have been able to obtain this information?
Why did they take more samples and focus most of their attention on the samples that were taken from the red springs instead of the green colored ones?
Are they saying that since the biofilms have already been exposed to variations in the environment and have already been introduced to sulfide and hydrogen, that they did not respond drastically to the addition of sulfide and hydrogen that they added?
Why does the Persian Gulf have such a high annual oil contamination? Has this increased or decreased recently? Are there any other places that have higher numbers of oil contamination?
Since the bacteria have different decomposing enzymes, is it possible to combine multiple bioremediation methods to degrade PAHs with higher molecular weights? If these methods were combined, is it possible that they could have an inverse effect & end up doing more harm than good?
Since PAH bioremediation is effective and environmentally benign, are they being done on large scales? Are the residents of the area accepting of the bioremediation, or are they doubtful and afraid of the science behind the treatment?
Were the seawater samples taken in the same area? Could differences of salinity of the seawater samples effect the number of naphthalene-degrading bacteria strains collected?
Although this form of bioremediation is helpful for marine ecosystems in the Persian Gulf, can it be helpful in more rural marine areas as well? Could the rural environment alter the type and number of naphthalene-degrading bacteria?
Although, Sphingomonas is a naturally metabolic bacteria that is very versatile and works well in bioremediation, some of its species can be dangerous for some organisms. It seems that its versatility could be a trade-off for safety.
Although Sphingomonos is versatile and biodegrades PAHs well, it is corrosive to copper pipes. This could undo the progress of the bioremediation and possibly effect neighboring water supplies.
GSTs can be used to detoxify xenobiotic cells, which is good in some organisms, but they have the chance of causing resistance. They can resistance in chemotherapy. Is it possible that they can form resistance in the biodegradation of phenanthrene and other PAHs as well?
I found it interesting that both the Ectothiorhodospira and the uncultured both had the highest identity percentages, although the Archean seems to be unidentified.
I can understand how PAH exposure through respiratory uptake can cause harmful effects. I’m curious whether or not the dermal uptake is present in the same situations where respiratory uptake is occurring. I would guess higher concentrations of PAH would be required for dermal uptake.
Are inorganic nutrients able to provide energy in order to power the metabolic pathways used to degrade PAH? This could be be accomplished by harvesting energy from redox reactions or possibly promoting an excess of reactants forcing the reaction to move forward.
It would be interesting to see if the bacteria of interest in this study could be supplemented by certain nutrients to target Phe instead of Fla. After reading the entire study, it seems like this could be a possibility.
I wonder if pH 7 is the optimal growing condition for most of the cultures used throughout the experiment or if it was selected for simplicity purposes. Keeping it consistent throughout the experiment is a good standard but I wonder how it affects the growth of various bacterias.
Extractions were commonly done throughout organic lab. It’s interesting to learn about an example in which this lab technique is relevant to microbiology!
This experiment seemed to be a little bit of a side track. Was the purpose of this experiment just to compare how this bacteria degrades ILCO as opposed to other strains that had already been studied.
It’s very interesting how the rate of degradation is not linear. I would have expected a linear line or even a line demonstrating exponential decay. There’s also a fairly large standard deviation on the R. erythropolis in MSM for a few values that may distort the line.
It’s very cool in this experiment that two methods of identification were used – one chemical and one biological. Certain functional groups such as an aromatic were identified through the degradation of ILCO and then the genes of the bacteria of interest were sequenced to find similarities to known genes.
It is quite interesting how the expected outcome and observed outcome of this particular experiment differed. The method on this section seemed to place a focus on intending to observe the degradation of aromatics in the Arabian light crude oil, although the results explained that only aliphatic components of the oil were degraded.
I agree that more research should be done before implementing Rhodococcus sp. CMGCZ as an environmental PAH degrader. It seemed as though some of the observed results from this experiment did not match the expected results, which may mean there is more to be considered in regards to PAHs and degraders.
I wonder where there obligate marine bacteria reside mainly. It would be interesting to know their exact locations – such as being coastal or in deep waters.
I would be interesting to know how the abundance of Cycloclasticus either increased, decreased, or stayed the same following the oil spill in the Gulf. This bacterium seems very relevant to this area.
Is it possible that PAHs are more plentiful relative to other carbon energy sources in marine environments so bacterial strains have adapted to utilizing this resource more?
orf7 was the open reading frame that was read in a different directions from the other orfs. Is it possible that this is an anomaly considering no function or obvious homolog was found?
I think it’s really neat that these genes can be introduced into E. coli as a way to better study the importance of the gene. I’m sure E. coli has been studied so extensively that it’s quite easy to observe differences.
I’m very interested to read about why they could not find the PCR products for arsenite oxidase genes even though this specific oxidation activity was observed.
I think this is a very cool experiment, tagging specific elemental oxidation states. I did not know this experiment type would allow for different oxidation states to be known.
I wonder if the anaerobic in the dark has a broader temperature range because on environmental adaptation. In dark locations, it could be very cold or extremely hot if near a thermal vent. I would expect light conditions near the surface to be much more consistent in temperature.
Acetate was not utilized in the reduction of As (V) because that reaction occurs in the dark and results from chemoheterotrophy. The difference in energy sources results in different molecules being utilized in the respective redox pathways.
I think it’s pretty neat that although acetate itself couldn’t be utilized in the reduction of As (V) its byproduct, CO2, could be so radio labels from acetate are visible in As (V).
You made a very good point. Would the bioremediation of Rhodocuccus for PAH degradation purposes have unintended or potentially harmful side effects on different aspects of the environment? I did not think about that while reading this experiment, but it seems necessary to answer your question before proceeding to introduce degraders into the environment.
I’m curious about how the researchers figured out the composition of the hot spring water in order to mimic the environment. It seems like the chemical composition could possibly shift due to certain environmental effects and not be entirely consistent.
Jesse,
I also thought this part of the materials and methods was interesting. I’m curious if the researches dismissed these two clones because they suspected issues or if there is any possibility that studying these two clones could lead to possible new discoveries.
I know you can do mutagenesis to enhance the compatability for transformation and expression using E. coli. They may have found some problems with all but ArhA1A2.
In paragraph 4 it mentions that multiple clusters can be found within the genome of some sphingomonads. They seem to wanting to find out how these clusters work if the genes aren’t being regulated within the same operon.
I remember that mutagenesis can be done to aid transformation and expression in a host. ArhA1A2 may be easily transformed while the others need to be mutated first to be accepted by E. coli
By paragraph 4, It seems as if the approach is to find out how the clusters come together to work on the same pathway when the genes are not coordinated through the same operons.
Could they have also done mutants without ArhA1A2, and also take out ArhR to see if its effects can be seen on the A4 electron transport proteins? The same could be done without the ArhA3A4 genes instead of A1A2 to assess which genes it possibly inhibits or induces.
I know that cosmids are able to utilize the cos site as sticky ends like a phage vector, but the extra phage genes are gone. Transduction happens really easily with this type of vector so maybe that plays a part.
Would this be an important find because all of the genes studied are downstream of ArhA3 and upstream of ORF6? Would this suggest that these genes are all required to function as a cluster and not be located seperately?
It seems as if AG3-69 doesnt grow on acenaphthene or its intermediate 1,8,-NA suggesting that the mutagenesis carried out removed the strains ability to oxygenate acenaphthene.
The marine environment is a huge dump for pollutants, I would assume that by now these organisms probably have many acquired genes for degradation of PAHs and would prove to be an asset to studying mechanism pathways of multiple degrading genes.
Are the bacteria being studied from the family Rhodobacteraceae too? I’m unsure if they are or if they were just pointing out another organism who displays flouranthene degradation.
Would the genes being located close by have anything to do with how many genes they are finding? It seems they have located way more genes than the previous papers.
Are these metabolites produced during degradation of a specific PAH or are they just providing a list of metabolites produced as a whole, rather than for flouranthene degradarion alone?
Could they have also checked for the lipoproteins and glycolipid’s mentioned in paragraph 15? They could have assessed whether they are constitutive or based on response to fluoranthene.
Do we know if B30 can degrade fluoranthene? If the genes which aren’t shared with B30 are important for degradation, could they just enhance the ability of the B30 genes found in P73?
I think a microarray could be a convenient way to assess which of the many genes they have found are produced under an environment containing solely fluoranthene as a carbon and energy source.
When compared to the other studies it seems this one gathered more information with the methods they used. The way this organisms PAH degradation genes were more closely organized seems to have benefitted the results of this paper.
“However because siderophores are released from cells, they are considered public goods and are susceptible to exploitation by non-producing cells”
Vibrios bacterial species utilize cooperative behavior. An environment with limited resources affects cooperative behavior by increasing costs for the producing cell, because resources are released away from growth towards cooperative functions as public goods. Siderophore production qualifies as a cooperative benefit. Many species are siderophore non-producers but have the genes to bind and utilize it. So these “cheaters” benefit from the cooperative actions of others without suffering the cost and energy themselves.
A gene knockout will cause the DNA to mutate in such a way which prevents the expression of a specific gene. Here, Vibro chromosomal mutants are generated by introducing plasmids incorporating the V. Fisheri genes iutA with fhuCDB. Assumingly, these genes enable V. fischeri to prevent the growth of other vibrio species including V. harveyi.
I researched the co-culture competition between V. fischeri and V. harveyi, under iron-depleted conditions. Aerobactin, a bacterial iron chelating agent production allows V. fischeri to competitively exclude V. harveyi— which does not have aerobactin production and uptake genes. In contrast, V. fischeri mutants incapable of aerobactin production lose in competition with V. harveyi.
And introduction of these genes are sufficient to convert V. harveyi into an aerobactin cheater.
What is the next step if no naphthalene degrading bacteria can thrive in a highly selective environment? Would alternate bioremediation strategies need to be considered?
How does this fact translate logistically for biodegradation of oil and pollutants? Would strain N7 be the best and/or most efficient biodegrading bacteria?
Since the gene coding for the most important member of meta-cleavage pathways is located in plasmid, how difficult would it be to bio-engineer non-PAH-degrading bacteria to be PAH-degrading?
Was this strain anticipated to be a good model organism because of the characterization of the catechol meta-pathway operon genes and their neighboring glutathione-S-transferase gene?
Why is identifying the different degrading bacteria strains and their efficiency important? Based on which bacteria is the most efficient, would environmentalists introduce those strains into environments that need more bioremediation than what was naturally taking place?
I looked it up, and the way I understand it is Biolog-GN tests are a way to determine bacterial strains’ ability to grow with different carbon sources. Is this right?
I don’t remember reading it, and I looked back and didn’t see anything in the Materials and Methods. Is it mentioned in the paper how they determined the fatty acid composition? Would gas chromatography be used?
I read online that GST was a class of molecules best known for detoxification. Does this mean that GST is directly responsible for PAH-degrading bacteria’s ability to be used for bioremediation?
I looked back at paper 1. At 200 ppm naphthalene, the optimal strain had 89.94% naphthalene degradation, and at 100 mg l-1 phenanthrene, this strain had 98.74% phenanthrene degradation.
The fact that they mentioned this strain could degrade phenanthrene “thoroughly” made me wonder if it is possible that other degrading bacteria only partially degrade phenanthrene.
Why would they have not been able to obtain authentic PCR products for arsenite oxidase genes? Were they not able to design a primer suitable to these genes?
This question was answered this week in lab. When this paper was written, the GenBank only had a few sequences to design a primer. The primer designed was probably not suitable for these particular genes.
What does this mean that these clones failed to show significant similarity to anything in the GenBank database? Does this mean that no one has annotated these genes yet?
The percentage is above the baseline number given with no additions; therefore, assimilation using acetate was stimulated by the addition of As(V) and As(V)+H2. It was not stimulated by the addition of H2,killed. The number for the addition of H2 is too close to the baseline number to give a definite yes or no.
So in the light, As(III) acts as the electron donor (what water would do in aerobic photosynthesis) in anoxygenic photosynthesis, and in the dark, As(V) acts as the terminal electron acceptor in anaerobic respiration (how oxygen would act in aerobic respiration)?
I think you answered this question in lab today, but I cannot remember what you said, and I didn’t write it down. In figure 4, why was there a slight reduction of As(V) when no electron donor was added?
Why would this strain possess genes for chemotaxis, yet be non-motile/not possess genes for flagellar proteins? Why have they not lost the genes for chemotaxis?
So for this experiment, the amount of PHE degraded should decrease as the concentration of Cu(II) is increased because the copper hinders the activity of the degradation enzymes. Also, will they test each initial concentration of PHE with each concentration of Cu(II)?
From what I understand, they’re interested in the effect of copper because the PHE needs to be degraded because it is a pollutant; however, heavy metals (like copper) can hinder the activity of the enzymes necessary.
I googled a different comparison of cluster structure, and in that one, the length of the arrows corresponded with the length of the ORFs and the direction corresponded with the direction in which it was read.
The way I’m understanding this paragraph, degradation enzymes have been located and described for a fluorene-degrader; however, it’s gram-positive. Fluorene-oxidizing enzymes have been located and described for gram-negative degrader; however, the hosts doesn’t grow when fluorene is the sole carbon/energy source. This paper will be the first that locates and describes fluorene-degrading enzymes for gram-negative degrader that can grow when fluorene is the sole carbon/energy source.
So for this overexpression experiment, the dioxygenase genes (flnA1-flnA2) were cloned into the overexpression plasmids from paragraph 6, allowed to grow, and then the products from that were extracted and measured using GC as in paragraph 9?
A follow-up to my last question: is the SDS-PAGE also a step in this experiment? Do they perform SDS-PAGE before GC? I get a little confused with the methods not being in order of the experiments.
They performed RT-PCR of flnA1 and flnA2 using cloning vectors to see if they were transcribed more in strain LB126 when grown on fluoranthene and not when grown on glucose. This was the case. They then used expression vectors to see the enzymatic activity of these same genes.
Phenanthrene is one of the preferred substrates; however, it cannot be used as a sole source of carbon and energy by strain LB126. Could this be because, like you mentioned in one of your comments in the introduction, the initial enzyme has the relaxed substrate specificity to accommodate phenanthrene, but the later enzymes in the pathway do not?
This paper is looking at the degradation of carbofuran even though several metabolites cause effects on the reproductive system in female rats. Is this because biodegradation is the only way to eliminate carbofuran even if there are negative repercussions?
Sphingomonad strains seem to appear in papers for the degradation of many different substrates. Is this because Sphingomonas is just more capable (than other bacteria) of degrading a wide variety or is it because this is just what is isolated when researchers collect samples?
The gene that when interrupted resulted in the loss of Carbofuran degradation was identified, and then sequenced to design primers in order to perform RT-PCR to confirm function?
I would think this result would be a surprise. All of the degradative pathways we’ve looked at so far in these papers have been inducible if I’m remembering right. Usually glucose would be the preferred C source and then the alternate pathway would be induced when glucose was used up.
The mutants for the most part exhibited reduced or slower mineralization rather than the abolition of mineralization. Is this because the pathway was shown to be constitutive?
So they selected mutants and assigned them to five groups based on whether how they grew, degraded, and/or mineralized carbofuran; they then selected ten of those mutants and verified whether carbofuran phenol (what they think is the first metabolite of degradation) is detected during the process?
So this paragraph is saying that although they used plasposon mutagenesis to identify genes responsible for carbofuran degradation, they had to be careful assigning loss of phenotype to specific genes because the loss of phenotype could be a result of polar effects instead?
Group I: C1 metabolism genes affected -> C1 metabolism is predicted to be related to conversion of methylamine -> hydrolytic release of methylamine moiety is first step in carbofuran-degradation
So for group I, the delayed/decreased growth rate on carbofuran is explained by C1 metabolism genes being affected, which in turn affects the first step of degradation, or am I misunderstanding?
I’m very interested to see specifically how they collected the bacteria and recreated an environment similar to their habitat. If the bacteria proves to be successful, how long will it take before they can actually start using this bacteria in the wild?
I figured that the oil eventually floats to the bottom of the sea floor, but I always wondered how it impacted the organisms down there since it’s already a pretty extreme environment. If bacteria is used for bioremediation here, would the goal be to remove all of the oil or just decrease the concentration?
These methods are super dense, but I love that I can recognize the primers and semi-understand the processes. Why did they use “near complete 16S rRNA gene sequences,” I feel like it would make more sense to use fully complete sequences?
I’m confused. I thought only Eukaryotes have mitochondrial respiration and Bacteria has respiration through cell membrane. If Naphthalene hinders mitochondrial respiration, it would not do anything for the bacteria, right?
If we were to use one of the common naphthalene degrading bacteria against a PAH compound that has not been well characterized and its not able to compete with the bacteria that’s polluting the water and ecosystem, will the selective pressure make the bacteria that we are trying to fight worse and even harder to get rid of?
Since the N1 and N2 strains has the most growth, does this mean those are the two stains that are causing the most pollution in the Persian Gulf? If so, would bioremediation of these pollutants significantly reduce the pollution in the water?
I was looking at the phylogenetic tree tables, and I was wondering if the originator of all the stains was targeted with the bioremediation approach would all the strains that followed be eliminated as well?
Since the different genera produce biosurfactant and bioemulsifiers that can uptake and dissolve the component of the crude oil that is contaminating the Persian Gulf already, is there a need to anything else? It seems as if the ecosystem is fixing itself.
It seems like they should focus on the obligate marine bacteria in coastal marine environment versus terrestrial habitats since this bacteria has only partially been studied. They are studying something that has already been extensively characterized.
I don’t feel like the author really told us the importance of their study as opposed to all the other studies on this topic. What will this paper tell us that the others didn’t ?
Since they did not have any small subunits to make their dendrograms, did they just use the previously published amino acid sequences based of the large subunits found in their experiment?
How did they come to the conclusion that As (III) was oxidized due to anoxygenic photosynthesis? They should not have been able to conclude this just from this one figure. Couldn’t the oxidation be due to chemolithtrophy since it can occur in the light or darkness?
So does this mean that As(III) will continue to reduce due to the microbes losing their ability to oxidize and not due to the microbes being incubated in the dark?
Why do the 16s genes have to go through PCR amplification in order to be identified? If all PCR amplification does is make more copies of the gene, why can’t the single gene be identified?
Why did the samples include both contaminated sea water and marine sediment ? If the sea water and the sediment was collected at the same location wouldn’t it have the same type of bacteria? Why the need for both?
Why were they unable to obtain authentic PCR products for the gene even though they observed aerobic oxidation activity? Will they elaborate more later on why they were unable to obtain PCR products?
Since the Ectothiorhodospira bacteria is dominant in the biofilm, does this mean this bacteria has the best resistance to arsenic opposed to other bacteria ?
What does PAHs stand for? Do these compounds referred to in this paragraph still pollute the environment badly in the U.S. because of sewage waste water and home sewage?
How much cleanup has bioremediation done since the oil pollution during the Persian Gulf War? How affective is PAH bioremediation really since there is already so much damage?
I know that in lab we pour agar. Do you turn solid agar into liquid for GC purposes? How would you turn solid agar into a liquid to extract the different compounds mixed in it?
Why were 18 out of 54 strains isolated? What happened to the other strains that made them not have an adequate growth rate? Could some of these strains be cross-contaminated?
What is a shaker incubator? Why wasn’t the microbial growth and naphthalene biodegradation monitored every day instead of just 15 days after? What made the experimenters choose 15 days of growth and then monitoring?
It would be interesting to read about why researchers think that Gram-positive bacteria might play a role in naphthalene degradation in the highly variable environment of oil-contaminated sediments. What brought them to these conclusions?
Was phenanthrene-degrading bacteria chosen to study because is an aromatic compound? Are we going to discover/read if naphthalene-degrading bacteria are better or worse than phenanthrene-degrading bacteria?
Was there only one isolate that was found that would be able to degrade phenanthrene? This is different from the last article because those researchers found several strains of bacteria capable of degrading naphthalene. Does this possibly mean that phenanthrene is harder to degrade because there was only one isolate found?
In this paragraph, it says that strain ZX4 could be a potential strain used for bioremediation. What other types of tests and research would be need to know if this strain could be used for bioremediation rather than just having the potential to be used?
Should the initial step involving meta-cleavage of catechol have been catalyzed by PhnI, but it was done by PhnH instead? I am a little confused on the first statement.
Why did the rates not change after constant shaking of the samples? I would think that after they shook the samples to increase oxygen exchange then the rates would favor oxic conditions.
When this paragraph says, ” We recently proposed that the respiratory arsenate reductase of strain PHS-1, and presumably the very similar sequence found in the biofilms, functions in vivo as a de facto arsenite oxidase”, what does “in vivo as a defacto arsenite oxidase” mean?
Will the researchers continue to search out this unexpected result? Will they research out about why there was an observed aerobic As(III) oxidation carried out by a novel mechanism? Will they see what this “novel mechanism” is?
I am curious to know the effect of more complex PAH’s on certain bacteria. Would they be able to efficiently eliminate environmental pollutants or would the by-products be malignant to the environment and people?
Why is it necessary to transport the seawater on ice when the samples were gathered from just 15cm below the surface? I feel as though the ice would bring the temperature above normal conditions therefore having effects on the organisms that live within it.
What factors play a role in the reason for the high diversity of naphthalene-degrading bacteria in the Persian Gulf besides the pollutant. If this is the only reason, could the same experiment be done in a different part of the world that has a similar pollutant?
I am curious to know what the optimum concentration for naphthalene-degradation would be for isolates found in different areas besides this specific area in the Persian Gulf.
What do they mean when they say the catechol 2,3-dioxygenase mirrors the taxonomic grouping of the host bacteria? Does this mean that it possesses similar characteristics of the host bacteria?
What is the purpose of amplifying the 16S rDNA? I see that you are basically breaking the strain down, building it back up again, and I think making it longer? But what purpose does that serve?
Why use the supernatant for enzyme assay rather than disposing of the supernatant and using a different enzyme? Does this result in different enzymes being assayed?
I found that the Sphingomonas paucimobilis strain is commonly found in hospital equipment such as vents and can cause disease. I think it is interesting that a strain with these characteristics also possesses positive characteristics in the sense that it is able to degrade phenanthrene which is found in cigarette smoke.
Is the pH range for this strain considered a wide range or no? I would assume that a 2.5 difference would be considered a wide range. This should allow for a greater variety of conditions that this strain is able to grow and survive in compared to other strains.
I think its extremely interesting that this ZX4 strain offers an environmentally friendly alternative to creating indigo instead of having to harvest it from plants. Another positive capability of this strain is its ability to be used in bioremediation. Two environmentally friendly uses.
I looked up what xenobiotic compounds were and have concluded that the ability for the ZX4 strain to degrade these compounds is yet another environmentally friendly characteristic that this strain possess. What are some environments that xenobiotic compounds can be found in? How can this strain be introduced in those environments in order to rid the environments of those pollutants and how long will it take for them to be completely eliminated?
Did the researchers decide to focus on examination of the cycling of arsenic under anoxic conditions because of how the coculture responded in the manipulated laboratory conditions?
How did they keep track of the tubes that were shaken before they were inoculated and the ones that were not? Is there any specific reason for shaken some of them and not shaking others. I am curious to see the results of the tubes that were not shaken and and how they may differ from the tubes that were.
How could the authors know for sure whether the capacity for aerobic Arsenite oxidation is due to the shallowness of the pond or if it is another reason?
Do all regions have unique PAH-degradation patterns or is only the Nakheel region unique from the others? I would assume that if each region has a unique pattern, then removing PAH’s completely would become an extremely difficult task.
They mention how the concentration of the PAH is important for removal. Are the co-substrates like yeast used for this purpose, or how do they go about lowering the concentration of the PAH?
I am a bit confused by this paragraph as to why they are testing the sediment samples ability to withstand the stress. What does it mean about the sample if it is not able to do so?
What is the advantage of constructing the phylogenetic trees? Could they use information from them to determine similar degradation methods that have already been determined, or would this not be possible with the unique PAH samples they have found?
If bio-emulsifiers “act like surfactants” and the water solubility of LMW PAHs is improved when surfactant-production is present, could these bio-emulsifiers ever be modified to be useful for both LMW and HMW PAHs?
I know they mention that there is no significant difference among the 3 sites, but table 1 shows that there is a slight difference in temperature at all 3 sites. Does this have any impact on the concentrations of the different nutrients found at each site?
I think it is pretty interesting how the table shows that none of the isolates showed ‘excellent growth’ when it came to a PAH substrate. They were all ‘good’ or ‘poor.’ Could that mean there are much better strains to use for bioremediation?
If all of the isolates were either at 100% similarity or very close, then why would they not look at different sequences aside from the partial ones they did? I guess I am wondering how they are able to draw any unique conclusions here if all the results are almost identical.
My question is a bit related to both papers, so if hydrocarbon contaminants can be toxic in the waters, wouldn’t evolution have created better mechanisms within microorganisms for treating them? Or has not enough time passed yet because most of these pollutants are more recent and man-made?
It says that the cores were stored for subsequent use within 2 weeks. Does the storage time have any effect on how the following experiments on the core go? Or would the results be the same as one day later or two weeks later?
16s rRNA sequencing has been in both of the papers we have read. Is is just a common thing in general for sequencing or just for use in studying types of microorganisms/PAHs in these types of marine environments?
All of the genera in this paper are different from the last paper. I just find it kind of interesting how there seems to be a lot of species with potential of being PAH-degraders but it is still a huge problem in the environment.
I am a bit confused on obligate/non-obligate. I know what those mean in general but does it mean here that the obligate PAHs only responsibility is to degrade them while non-obligates have other uses in addition to degrading?
What exactly is a singleton sequence and why did it mean that those two weren’t further analyzed? I tried to google it but all the answers were a bit confusing.
I am a bit confused because the previous paragraph says only two reductase genes have been characterized but this paragraph talks about a separate one, or am I reading this incorrectly?
I am a bit confused here. So they are trying to screen the E. cloacae library and they need a strain that doesn’t have selenate reductase activity? Why is that?
After reading the introductory paragraph, it’s clear that individuals exposed to PAHs mainly suffer from symptoms relating to the respiratory system. My question would be which type of exposure yields worse symptoms, dermal uptake or respiratory uptake? And what are the various types of preventive measures that have been put in place to avoid exposure to PAHs?
The article briefly mentions how both LMW and HMW microorganisms are used as a means of bioremediation against PAHs. However, does one prove to be more efficient than the other? Is the reason LMW microorganisms are more commonly used in studies because they’re more easily accessible or because they’re the better option of the two?
In paragraph 2.3, the procedures detail how they let colonies grow on MSM plates for approximately two weeks before the isolating and sequencing the cultured colonies. However, in paragraph 2.4 the procedures mention how they only let the naphthalene degrading bacterium grow on MSM plates for 8 days. Why weren’t the plates given 2 weeks to grow? Did they not need as much time to grow?
For the MSM plates, why were the percentage degraded for Nap and Phe so close compared to the Fla? Are the chemical structures for Nap and Phe somewhat similar? And if that’s the case, shouldn’t they also have similar percentages degraded when placed in the YMSM plates?
Given that the degradation percentage of crude oil was so low, would it make sense to search for an alternate degrading bacteria? Or are these results typical to what you’d expect when attempting to degrade crude oil via bioremediation over such a short period of time?
It’s definitely interesting that CMGCZ was better able to degrade Fla more efficiently than a structure like Napthalene. Especially considering Napthalene is only a two ring PAH while Fla has four rings.
I accidentally compared Fla and Cycloclasticus.. when Fla is a PAH and Cycloclasticus is a degrading bacterium. So my original post should’ve asked if Cycloclasticus is a better bacterium at degrading Fla than the original bacterium we studied, Rhodocuccus.
Of the PAHs being studied, I wonder which ones are of a low molecular weight and which are of a high molecular weight. Which will be easier for the Cycloclasticus strain to degrade? Does it have anything to do with the marine environment?
The LB agar plates are the same used from the first experiment we read about for Blog Summary 1. To me it seems to be a standard growth plate used in labs everywhere.
After reading the introduction of the paper, I thought that the mission of the experiment was similar to the first blog we read; therefore leading to similar experimental methods. However, the further I read into this paper, the more complex this experiment seems. I’m interested to see how the results vary between the two experiments.
I wonder what the specific location of the 10.5-kb SauAI fragment was. And if it was in identical locations in both the pH1a and pH1b constructs. In Experimental Designs with Kroetz, we learned that the location of a specific sequence was key in determining its characteristics/influence on the DNA.
Does this paragraph hint at the idea that there’s a specific sequence of DNA that gives microorganisms the ability to degrade PAHs? It would seem to be the case considered there were 7 cases of PAH-degrading constructs found with similar if not the same sequence.
Why were the chemical structures from Fig 4.4 converted to a different form? What was the purpose, especially considering the form they were transformed to were stated to be more structurally unstable.
So does this hint at the chance that having a shared dioxygenase system is a contributing factor of the increased ability of a strain to degrade a PAH?
Does the ability to degrade PAHs go up as more dioxygenase subunit genes are found in a strain? That’s what this paragraph suggests, yet you would think that the attachment of additional DNA would have the opposite effect.
I wonder if a bacteria’s ability to metabolize substances that are normally considered toxic to organisms means that its categorized as an extremophile. This would also make sense because the paper discusses how some bacteria with this ability live in extreme environments such as hot springs.
I wonder if the inability to obtain PCR products for arsenite genes has anything to do with the specimen being collected from an extreme condition (i.e. a hotspring). Why weren’t samples from somewhere else used and would they have likely yielded the same results? Interested to finding out what the next step in their experimentation was.
I honestly was wondering the same thing Justin. I wonder if the details given about the location were significant in a way that isn’t obvious to a reader. Because to me, the details seemed pretty generic.
Why was there no type of pattern in the intervals that the samples were taken? It seems odd to me that they retrieved 2 samples in 2008 only 2 months apart and waited 6 months to retrieve another one in 2009.
It’s interesting reading about how exactly the researchers set up and prepared their experiment. It seems as if they’re was plenty of room for human error. Which may explain the inability to produce authentic PCR products for the arsenite oxidase gene later on in the experiment. I wonder if there is any correlation.
I wonder if there are any major similarities between the bacteria strain compared to the archaean strains. Because to me it seems like the bacterial strain would have to have some of the same properties in order to maintain survival in an environment typically known to be home to extremophiles (aka archaea).
It’s cool seeing how the slurries were able to go back and forth between oxidizing and reducing arsenic based on their environmental conditions. I wonder if the amount of exposure to either light or darkness affected the speed of oxidation/reduction.
This paragraph answers my question that I wrote under paragraph 5. I’m curious as to why the temperature range for reduction is so much bigger than that of oxidation.
How can it be determined that acetate was used as an electron donor when in the paragraph above it’s stated that there wasn’t any observation of As(V) reduction with acetate?
This paragraph is important if the reader has no background knowledge of what PAH is or the dangers of it to the environment. It not only breaks down the toxicity of PAH, but also discusses why the chemical is so hard to remove.
This paragraph highlights the overall purpose/goal of the experiment. In the previous paragraph, it is noted that there are bacterial strains capable of surviving in the toxic environment of the PAH stricken Arabian Gulf. The goal of this study is to isolate the bacterial strains growing in the PAH environment and further study their functionalities. A better understanding of these bacteria would help future bioremediation efforts across the world.
I noticed that the researchers used 16S rRNA for constructuing the evolutionary relationships of these bacteria. My question for this portion is; is sequencing the 16S rRNA more effective than the 23S and the 5S rRNA segments?
What makes the temperature of a particular sample site important? Was this to ensure that the sample area could sustain the PAH degrading microorganisms or is it something more technical?
What is the reasoning behind the difference in degradation abilities of the same organism on different PAH compounds? Are the compounds so unlike one another that certain bacteria will specialize into a particular PAH compound, rather than PAH’s as a whole?
The PAH isolating bacteria isolated from soil on the surface would have little to no effect on oil degradation at such low depths, could this class of organism be understudied due to the expenses involved in isolating it?
Since these areas are ridden with hydrothermal plumes, could one expect to also search and isolate archaea PAH degrading organisms, as their extremophilic nature would make them a dominating organism in the ecosystem?
Would an PAH degrading archaeon that could sustain higher temperatures preform better the closer one got to the hydrothermal center? Could a selection of two microorganisms capable of degrading PAH, one being a dominant PAH degrading bacteria and another being a more niche archaeon work together since they would occupy separate spaces along the seafloor.
Could Cycloclasticus have an impact on other deep sea enviroments with abundant sediment surface hydrocarbons? Would Cycloclasticus preform better if it was deposited alongside Halomonas, Thalassospira, and Lutibacterium in a new enviroment?
Is this “rusty-yellowish” coloration able to be observed at the site of extraction? It would be interesting to compare the coloration and presence of oxidized intermediates in the original location and the studied organisms.
The prosthetic heme group acts as an Oxygen binding site in hemoglobin of animal cells. Why would an organism utilizing anerobic metabolism of Se require the capture of Oxygen?
Will the selenium metabolic mechanism always produce a similar mineral product, or will a selenium metabolic mechanism from a different organism produce a different mineral product?
Genome sequencing was clearly important in order to identify the presence of known selenium metabolic genes, but would a transcriptomic/proteomic approach be useful in this experiment when searching if an organism contains rna/enzymes capable of metabolizing selenium?
What would be the proper procedural steps to take if the organisms were capable of selenium metabolism, but none of the three strains contained the gene that they were searching for?
When looking at the graph, it is notable that pLAFR3 expresses a relatively constant Se (VI) Reduction, whereas the reduction rate of pECL1e gradually decreases over time.
This experiment was preformed to confirm that the fnr gene is essential for the reduction of Se (VI) to Se(0) in E. cloacae SLD1a-1. The results from this experiment have shown the importance of the fnr gene.
The FNR gene is essential in the detection of Oxygen concentrations, but is also essential to initiation the degradation of Se(VI). Without FNR, no Se(0) precipitation will occur.
If there was a way to mutate FNR to make it sense another molecule OTHER than oxygen (i.e. mutating the gene so that it detected the presence of nitrogen), could you make a pathway that was dependent on the presence of another gas? Are there other similar pathways in nature that are dependent on such gas concentrations.
Apologizing for all the questions in advance, this is just a fascinating concept.
Many questions arose when I first started reading this article in Paragraph 2. For starters, it states that a main concern to Antarctica’s natural region is being damaged by the increase in tourist and other people visiting the continent. When I googled how many people are annually visiting Antarctica I was shocked by the numbers. It states that when Antarctica first allowed for people to visit in the 1950’s only a couple hundred a year visited. Recently, more than 56,000 tourists visited Antarctica during the 2018-2019 season. That is a huge increase in only 60 years for a continent that is mainly an ice wasteland. I can completely see why native Antarctician’s are becoming more afraid of their climate due to the increase in visitors. My guess is that more people will result in warmer areas and increasing the use of various fossil fuels to bring people on and off the continent.
We probably can all agree that various Oil spills and burning of fossil fuels has in fact caused the Earth’s atmosphere to become warmer and also resulting in Antarctica losing areas of ice each year. [ Oil contamination can generate detrimental changes in soil properties, including modifications in maximum surface temperature, pH, and carbon and nitrogen levels (Aislabie et al., 2004).] From this quote in the article, I was shocked to see so many different types of Proteobacteria being affected from oil contamination and my overall response to this is, well that cannot be good, and its not. From my understanding of paragraph 2, Antarctica are in real trouble when it comes to oil and other fossil fuel contaminations and if it is not improved, many of the bacteria essential for Antarctica’s nature could no longer exist and the entire world will feel its effects.
[53 cultures that metabolized phenanthrene were selected, and the concentration of PAH quantified as described below. The three highest metabolizing strains were then selected for further studies.]
It is very interesting to me to see how these scientists are creating 53 different cultures that will ultimately metabolize phenanthrene and their PAH concentration and pick upon the nest three with the highest metabolizing rate. I believe out of the 53 some might even be rather similar, and if you have multiple that are very similar, it must be quite challenging to select to best options to experiment on?
[Briefly, non-polar compounds were extracted from culture media using two volumes of hexane and vigorous mixing for 60s]
From the Phenanthrene quantification section in this article, i wondered why it was so important to use two volumes of hexane and vigorous mixing for 60s in order to extract the non-polar compounds. So when I browsed the internet trying to find an answer to my question, I quickly realized that the main reason behind it is to just clarify the non-polar compound you are trying to extract, while it also helps purify the substances being extracted. This is also why the procedure is repeated for a total of three times in order to make sure the non-polar compounds were fully and properly extracted.
The main part of paragraph 3 that stood out to me was the similarity in how the glucose in both the unexposed and the diesel-fuel exposed were realitivly the same. On another note, the other 2 aspects of the graph showed increases in CFU for the control group as well as the phenanthrene. To me this shows that an increase in phenanthrene is present in the diesel fuel sample.
[Analysis of optimal growth temperature revealed that all isolates exhibit maximum growth rate at 28°C (Figure 2C), indicating that these strains are psychrotolerant rather than psychrophylic, a behavior we have seen in previously isolated Antarctic strains]
The main question that I derived from paragraph 4 was that why were all of the samples experience maximum growth at 28 degrees Celsius? It is just strange to me to think that the variety of different samples all hit their maximum growth rate at the same temperature rate. As stated in the paragraph as well, this means that the strains are psychrotolerant from our previous examples from the Antarctica stains.
One thing that stood out to me in this paragraph was the fact that Antarctica is a certain region where bringing over foreign organisms is forbidden in the continent. Like the article continues to say, this increases the use of native bacteria samples and also will help narrow down decontamination techniques and strategies. This is something I never knew was true and can fully understand and see where Antarctica are coming from in the fact that they don’t want to cause anymore unwanted bacteria to grow.
To respond to your question Rachel, I agree with you on the point that they were just trying to achieve their main goal, but I also believe because there are so many different varieties of bacteria in the area they are testing, it is more likely that they will not have a completely isolated degrader.
After reading this paragraph, the statement about the variety of competitive strategies intrigued me the most. From raid culture of limiting resources, to contact-dependent delivery of other toxic, can ultimately effect competitive cells to change.
I was confused on the part when the writer explains how species can produce siderophores that will have a competitive advantage to other cells, but what eventually caught my eye was how when these cells are released they are public goods and are exploited by non-producing cells. I found this very interesting and believe to be very important for the upcoming experiment.
There is so much going on in this paragraph it is difficult to grasp what the author is trying to present. Other than all the equipment and type of bacteria’s/ antibiotics. What was the point of so many different samples that had to be added? What was the main advantage for doing so?
I found it interesting that the scientists did not filter the remaining mutant just for simplification purposes, but also how come ploymyxin B was added to prevent any more growth. I’m curious to understand how this actually works.
[Finally, the presence of V. fischeri ES114 culture fluids prevented the growth but did not kill V. harveyi. While below the level of detection by OD600 measurements, a small increase in V. harveyi cell density could be detected by counting colony forming units (CFUs) (Fig. S2D)]
I found this sentence to be very interesting to me and caused me to form a question in my head. I was wondering why does this culture prevent the growth, but still does not kill the V. harveyi. Usually I would assume that if a process or anything is stopped growing by an outside force, majority of the time that product will eventually die off.
[It was curious that growth inhibition of V. harveyi occurred only when culture fluids were obtained from V. fischeri ES114 grown in minimal marine medium but not in rich medium (Fig. S2B). ]
I found this interesting as well, to the point that the inhibitions were only obtained from V. fischeri ES114 when grown in medium but not in rich. I believe this is because the medium was the minimal required for the product to actually grow, and too much medium like in rich, the product might not like and abundance of medium and could harm or damage the process.
[Alternatively, the genes encoding IutA and FhuCDB could be acquired by horizontal gene transfer. Horizontal transfer is known to have distributed aerobactin genes across multiple vibrio phylogenetic lineages ]
This was a statement that caught my eye and I thought was quite interesting. I did not realize that horizontal transfer is known to distribute aerobactin genes across multiple lineages. I believe that this is actually necessary for the given experiment, and without them, it could be hard to encode these genomes, like with siderophores for an example.
[It could be beneficial to repress iron uptake under oxidative stress because ferrous (Fe2+) iron reacts with hydrogen peroxide in the Fenton reaction to generate harmful hydroxyl radicals that damage DNA (Imlay et al., 1988).]
Another statement in this section that I found interesting was the fact that more iron uptake under oxidative stress can be better beneficial. This is very interesting in the fact that they could have been using this technique for a long time in the experiment, and also helps understand how some things are changed and processed in this experiment, that could eventually harm and damage the DNA they are sequencing.
The fact that nitrogen recycling is important for marine life and having a nitrate reservoir, is something that I did not know and found very intriguing. It is interesting to see how nitrification can play a role in promoting marine and nitrogen loss.
[Since then, similar sequences of crenarchaeal AMO gene subunit A (amoA) (69–99% amino acid homology) have been detected in various marine water columns and sediments, including the Black Sea (20)]
I wondered why are these sequences so similar? And also the black sea caught my eye as well and also wondered if this body of water had some extra resources containing nitrogen than other seas. Since there was in fact various water columns and sediments in the black sea.
I was interested in what the CTD looked like and how it performed, so after looking at the link Dr. Ni Chadhain, I had a more understanding of how the equipment worked. Overall, the picture helped me better understand what was going on in this paragraph, because I gained an idea of how the machine works.
I was interested in how a FISH actually worked and why the CARD-FISH was better to use than the other. After reading some more information about it and following the link attached above, I can see why they used this system because it does indeed work well will samples that have low abundance. The CARD-FISH is shown overall as an improvement when compared with the normal FISH mainly in aquatic environments.
In figure 2, after observing the charts, I had a hard understanding of them but after todays class where we had an example of this figure, I have a better understanding. Although Figure 2d was some what confusing but can be seen to show the relationship between a,b, and c.
I feel like this figure was easier to understand than the Figure 2, mainly because each color has a different label. From this, I believe that NO2- is probably the best source of vertical distribution in water, since its lines are so close to the edge of the chart. It is also good to know that O2 levels are very high near the surface, and of course very low as the water level drops.
From this graph it was similar to the others that we discussed. Since the oxygen gets lower as the water depth drops, this can help apply the theory that sulfur reducing bacteria will successfully live in. Nitrogen can also be shown in the water levels as well.
Its good to know that this was a vertical distribution of the amoA expression with the crenarchaea. The crenarchaea levels seem to be the greatest as the water level drops. Also this can be seen in the graph in figure 2 where it is better represented than betaAOB and gammaAOB.
This part of the introduction provides background information to the reader. It explains what PAHs are and how they’re harmful as well as a summary of how and which microorganisms break them down. However, the authors caveat by saying that there is much to PAH degradation that we still don’t understand, especially with regard to high MW variants.
It seems according to this paragraph that while we’re capable of sequencing the genomes of PAH-degrading bacteria, without a better understanding of the metabolic pathways that actually serve in breaking down PAHs, the knowledge of these bacterias’ genomes is of limited use to us in understanding the breakdown of PAHs.
I am somewhat curious as to how the sediment for C. indicus and C. baekdonensis were extracted from the ocean, as well as what depth they came from (even though it’s not exactly relevant to the article). I also tried looking up Ausubel et al. DNA extraction, but most of what I found were similar citations to it.
I looked more into GC-MS and it was pretty interesting. Apparently the SCAN mode, if I looked at the description correctly, is useful in determining unknown compounds and scanning ranges of mass fragments. I’m not entirely sure what it’s being used to determine in this paper, but I read that GC-MS can be used to determine metabolic activity, which I suppose could be useful in trying to isolate a pathway for PAH degradation.
So to see if I understand this figure correctly, this figure (along with paragraph 11) is showing the predicted phylogenetic tree containing P73 and its 5 plasmids present, and that the trees are based off comparing the protein sequences of the parA gene (for figure 2A) and the rep gene (for figure 2B) with other available genomes, and that the closest related bacterial species to those plasmids is where they might have originated from?
Interesting that the P73T strain has the genes for flagella assembly, motility, and chemotaxis and yet phenotypically demonstrates none of those traits. Does simply having those genes assist in increase bioavailability of PAHs, or is P73T perhaps relatively closely related to a strain that is both motile and capable of breaking down PAHs, and this strain for whatever reason lost the ability to assemble flagella or become motile?
So the way GC-MS can be used to determine metabolic pathways is that it can detect potential products and byproducts of degradation, like it did here for fluoranthene degradation?
So this pathway corresponds to the top pathway illustrated in Figure 5, the numbers above the arrows being the genes involved. I’m curious as to what ways you could experiment to see if you could confirm steps in the process, or perhaps that’s not necessary so long as the end result is fluoranthene degradation.
The last sentence about P73T being potentially useful in marine oil spill bioremediation is a pretty cool possibility. I would imagine many more experiments and studies involving it would have to be carried out before it come be used practically, I imagine.
So while the researchers found these 138 genes that may be involved in aromatic metabolism, it’s likely not explicit that all of these genes are used in PAH degradation or fluoranthene metabolism. Could some of these genes hae been acquired via lateral gene transfer but otherwise not utilized (or utilized in other aromatic metabolic pathways not related to this study)?
Considering the age of this paper, have other significant groups of microorganisms capable of degrading aromatic hydrocarbons been discovered or have been the subject of greater focus in recent years? Are sphingomonads still the intense subject for researching PAH-degradation that they were when this paper was written?
So strain LB126 is especially significant because not only have the researchers identified the genes for fluorene degredation in a gram-negative bacteria, but in one that can sustain itself only on fluorine? And that makes it more significant than the pCAR3 plasmid from strain KA1?
So in order to test the expression of the dioxygenase-coding gene, they essentially constructed a plasmid by isolating the gene using PCR, subcloned it into the expression vector, and inserted that vector into the BL21(DE3) strain of E. coli?
I would guess that the purpose of dioxygenase overexpression is to grow colonies of E. coli with the vector containing the gene that codes for dioxygenase that would be able to express the ability oxidize PAHs, which would demonstrate that gene’s role in the process. Could you use gene knockout of that gene to reach a similar conclusion, like as was done in the previous paper?
Since they had to incubate the E. coli at 42 degrees C rather than the initial 37, are the proteins coded by FlnA1-FlnA2 in the E. coli different from those coded by the same genes in the LB126 strain of Sphingomonas because they have different activation temperatures.
I’m not entirely sure what the significance of the truncated orf2 encoding for a transposase. It apparently suggests horizontal gene transfer, but in the context of the whole experiment, it’s unclear to me why that’s important (or is it just an interesting observation?).
If both FlnA1-FlnA2 and CARDO can make 1-hydro-1,1a-dihydroxy-9-fluorenone from from 9-hydroxyfluorine, why is it that FlnA1-FlnA2 can make it from fluorine and not CARDO? Does this imply FlnA1-FlnA2 has a greater substrate range pertaining to fluorine degradation?
I suppose a closer reading of this paragraph actually answers my question on paragraph 3 about the importance of the truncated transposase gene. Is it the transposase’s proximity that leads them to the conclusion that LB126 acquired its genes for fluorine degradation through lateral gene transfer, as the paper only says it was ‘upstream’ (are they likely in the same cluster or group or something like that)?
Is it common for strains of Sphingomonads or other genera of microorganisms to use only a select few carbon and energy sources? Even if not, I imagine strains like A4 and the strain from the previous paper are novel since you only have one or two sets of metabolic processes to make observations about and detect enzymes and genes for.
Is the lack of reporting for the identification of dioxygenase genes for these sphingomonads due to just the lack of interest in identifying them, or is there some sort of significant difficulty in identifying them. If it were the latter, would reports exist that mention the challenges of identifying those genes (and the lack of said reports would indicate that difficulty is not the reason for the lack of identification)?
I guess this was their negative control to see if disrupting the arhA1 gene in A4 would diminish its ability to utilize acenapththene and acenaphthylene. By “blunting”, does that refer to removing the overhangs from the sticky ends of the DNA?
Is this preparation similar to what we did in lab where we used the oxidation of indole to determine which colonies did not have our inserts? I would guess it would serve the same purpose in this experiment.
Seems like the process of disrupting the arhA1 gene was effective. What is the putative arhA1 homologue mentioned referring to though? Is it referring to the gene they used in homologous recombination with arhA1?
While the arhA1 homologue seems to still be intact in the mutant, according to the results of introducing the mutant strain to the acenaphthene environment the strain couldn’t degrade it. So it’s interesting to consider what this homologue is. I’m curious as to what follow up research could be done to better characterize it.
So is the interest in the intrinsic electron-transport protein for ArhA1A2 sort of looking forward to the next step in PAH degradation, or is it another part of the initial process that hasn’t been well studied at the time?
In terms of bioremediation, does the Sphingomonad genus’s ability to degrade xenobiotics function similarly to its ability to degrade contaminants in the environment. Are ingested PAHs considered xenobiotics?
Primer extension is apparently useful in mapping the 5′ ends of RNA sequences. I’m curious to see what purpose this serves in the paper, since we never did primer extension (and I don’t recall if or how much we touched on it during lectures).
I imagine it’s common to use this many strains and plasmids pertaining to a particular species of bacteria in these studies. It feels like in comparison the number of strains me and my group have worked with pales (3 initial strains, plasmid vectors, and a few others). Of course, this paper probably took at least months of studies to complete, so if anything this would be what our research project would look more like were time not a limiting factor.
It’s a little sad that we never had time to do anything with Southern hybridization beyond discussing how it works, seeing how ubiquitous it seems to be as a method in these papers.
AG2-45, 2-48. and 3-15 lost the ability to produce indigo, but also maintained their ability to grow on 1,8-NA. Is that expected, or something not relevant to the conclusions suggested by these observations.
So it would seem the ArhA3 and ArhA4 genes were what the researchers were looking for in terms of the electron-transport system. It seems that incubation increased in cells expressing A1A2A3 without A4 (although not as great as with all four), so they’re complimentary, but are not both required for increasing acenaphthene oxygenase activity.
The arhR gene, interestingly, seems like a sort of “pre-step” for acenaphthene degradation, as its expression seems to affect the transcription of the arhA genes, and its disruption was shown to negatively affect acenaphthene degradation. It also seems to be active separate from the arhA genes.
So does this loose operon structure indicate an area for further research, or is it not relevant to PAH degradation since it doesn’t seem to have anything to do with acenaphthene degradation? Could it have been a result of mutation or gene transfer potentially?
I remember one of the papers (the first, I think?) involved theorizing a potential metabolic pathway in the breakdown of certain PAHs. They could potential do the same for future experiments. I agree that it seems difficult to accomplish with how dispersed they are, but I imagine there are ways we’re not aware of.
So since the arhA1 homologue’s function wasn’t identified, I imagine that finding its purpose would be a good jumping-off point for future research. Could you make preliminary assumptions of its function due to its homology to arhA1?
It seems so odd to me that despite the importance of Sphingomonas in PAH degradation we still know so little about their deoxygenation genes. It’s still somewhat unclear to me how this study increases our body of knowledge on the subject, but so long as it serves as a base point for further research, I suppose it doesn’t have to that much.
I remember during the oil spill, how the oil being released in to the deep ocean was causing big blooms in the bacterial populations that consumed the oil. this made big areas of hypoxias water as the oxygen was used up. and how this was one of the most damaging secondary events from the spill.
Its good how Antarctica is for the most part being protected from possible invasive species so we can keep it’s research opportunities available. It is interesting to see how these local environments are providing solutions for the problems we have created and how we have to be very inventive with finding these solutions since external fixes aren’t allowed.
I can see how our labs mimic this in our isolation of our naphthalene degraders. as our bacteria is isolated to single colonies to obtain strains for farther study.
PCR can be a highly powerful tool in the study of genomes as it can let us obtain as many copies of a piece of DNA as needed. most of the current Covid-19 tests are run through PCR to get the viral Genome large enough that it can be easily detected. which is also why it can take a while as PCR has to cycle through temperature ranges to successfully anneal and replicate the genomic material.
PCR and its readily availability has allowed for all the growth in genetic research over the last few decades as you can now easily get your hands on the equipment, proteins, and primers for replicating almost any DNA strain you want. I has need to all this testing with recombinant DNA and Plasmids as they are now cheap to produce once your initial sample is synthesized.
I probably wont be the three same strains but other bacteria in those areas can probably degrade oil. Oil and produces made from it are now everywhere thanks to humans, but oil will naturally enter different environments as it seeps up through the ground.
Life is a truly amazingly resilient thing. you can find some form of life at almost every point on earth. they have found microbes in salt water pockets at the bottom of mineshafts in Johannesburg. we have even done studies of exposing bacteria to space onboard the international space station, and found that several survived with minimal damage after an extended exposure.
Its interesting how they are using a naturally occurring environment with a significant amount of hydrocarbons as a base examination to compare to the areas effected by the oil spill.
I see this group is also using 16S to identify and match what species of bacteria these are. but I don’t quite understand why they are using carbon 13 for this. is it just a way to mark the bacteria they could culture from the samples?
Its interesting that phenanthrene is the PAH that is mineralized the most. Especially with PHE being the PAH that was used in the last paper. Is there some reason, maybe with the origin of the genes that allow for the use of PAH, that make PHE a globally common PAH that can be broken down? maybe the proteins that allow for the breakdown were originally specialized to a different energy source that PHE manages to be the most similar to.
Its interesting that there are these specific genera that have fulfilled their niche so strongly that they are only able to get their energy from the breakdown of PAHs.
It will be interesting to see how they will test to find a coculture of bacteria. along with this how they verified if it is a symbiotic relationship, and how the bacteria handle these reactions
It is interesting that they couldn’t isolate the arsenite oxidase genes. is it possible that there is some temporary form the arsenite is being made into before it self oxidizes?
Since they all ready have primers for the genes dealing with the uses of arsenate and arsenite. I assume they are working off research that was already conducted before that had isolated the genes.
It is interesting the amount of sterility they are using to conduct these experiments. They are acting really carefully to prevent oxygen and sources of lab contaminates. The prevention of oxygen makes sense as the bacteria are supposed to be growing in a natural condition that lacks oxygen to use as the end electron receptor and are thus forced to use the arsenic.
Its interesting how few bacteria they found found in the samples and how only one close group photosynthetically oxidize arsenite. but this makes sense when you look at the graphs that the arsenite is being oxidized by phototrophic bacteria.
With the large rate differences between the oxidation and the reduction of As(III) and As(V). would it be safe to assume that in natural conditions, that the soil and surfaces in which the biofilm where collected from would primarily contain As(V) with very low amounts of As(III)
I wonder why there is such a focus on the salinities and the comparison to oil spills and leaks, is it that the two are linked or just that the risk is high in these regions of high salinity because of concentration of extraction sources or if equipment disrepair and misuse is the reason.
I wonder what they used as the initial cultivation media. Because the bacteria was found to degrade all of these hydrocarbons but you need a pretty specific initial media as to isolate the bacteria in the first place as too many added hydrocarbons and the media would just act as a general purpose media grabbing multiple different types of bacteria.
Okay, this answered my question from the first page, they used benzene as the carbon source in their selective media. As benzene is the primary structure for the different compounds in the BTEX group. It would make sense for what ever proteins the bacteria uses can also bind to the other compounds and break those down as an energy source as well.
Looks like they did the pretty standard practice of using Carbon 14 enriched versions of compounds you are trying to see the bacteria degrade. The amount of released Carbon 14 CO2 released can show the rate in which the bacteria is degrading the benzene.
Its interesting that the bacteria ceased to degrade benzene at 4 M NaCl, although 4 M NaCl is 233.8 g of NaCl in a liter of liquid. A this level of salt it is safe to assume that the bacteria began to shutdown its intake mechanism to help lower internal salt concentrations.
It is interesting that they didn’t manage to find all of the enzymes that would be used to breakdown these compounds. They believe that the enzymes should be known ones and not the possibly of unknown enzymes for the breakdown of these different compounds.
Its cool that they actually go through and show the actual breakdown steps for the benzene, toluene, and other hydrocarbons. It takes these different compounds and converts them to acetyl-CoA which it can then use in most metabolic functions.
This is looking a bit bit closer at the biochemical properties of the enzymes, they talk about the bacterial strain’s protein structure. The proteins are actively more soluble due to their abundance of negative charges. This helps the bacteria continue to function in high saline environments.
Fluorene is a know to be highly toxic to aquatic life. I found a document by the Minnesota department of health that talked about the exposure risk from fossil fuel contaminated areas and cigarettes’ smoke. They haven’t preformed wide scale testing but they believe it can build up in lake sediments.
This paper brings up that the fluorene oxidizing genes are on plasmids, this differs from the last paper where the genes were part of the bacterial chromosome.
Protein electrophoresis is harder than DNA electrophoresis as you need SDS in order to smooth over the charges to negative so they can be attracted to a single electrode so you can actually get coordinated movement of all the proteins in the same directions and at speeds equivalent to their size and not their charge.
Its interesting how they have the exact primers for these genes although this was one of the first times they were supposed to have been isolated for testing. There is no degenerate nucleotides in these sequences.
Its interesting how in figure 2, terrabacter sp. DBF63 had a regulatory gene in the reverse direction surrounded by Fluorene degrading genes in the forward dirrection..
Reverse transcriptase is such a powerful tool in our tool belt of techniques and enzymes for genetic research. We took an important part of one of humanities most infamous viruses and made it into a tool to try and improve peoples lives through the research avenues it has opened.
Its interesting that the Proteins produced, if they were FLnA1 and FLnA2, were not fully translated in their initial conditions, but when they were produced at 42C they got full sized proteins. I wonder what the physical/mechanistic reason for the translation to have stopped is?
I find it interesting that FlnA1 and FlnA2 could be a relatively novel way to break down fluorenone and other cyclic hydrocarbons, it would cool to see if the evolution of these genes could be determined to figure out how it came to be and function in this way.
Its interesting how this is a soil sample from deep sea sediment, this could mean that the ability to degrade these hydrocarbons comes from oil seep exposure.
cre-lox recombination is a very interesting way to inactivate genes. Although it is semi limited in its use as you need the recognized sequence for the enzyme to work.
Its interesting that this paper actually when and tested all the different sugars the bacteria could use. It looks like they are doing a full examination of this bacterial genes/
The inability of the knockout to use fluoranthene and naphthalene and the defectivity of the other hydrocarbons suggests that this was in fact the gene responsible.
I too have an interest in the differences between the inocula, or rather, what qualifies them as “very similar”. I can only infer that the differences and level of effectiveness will be further discussed in the remainder of the paper, for anyone reading would also pose the questions you did.
I too wondered why the bacteria were not isolated or investigated from this location. While this is the introduction and is meant to be brief, another sentence explaining the reasoning behind the absence of a study could have cleared up questions such as yours.
I understand the incubation temperature-wise of the culture, but am questioning the shaking of the samples. If it were surface level soil I would understand the need to recreate the chaos of an environment that is walked, run, jumped, etc. on but the article does not identify if the culture was taken from the surface or below. Why did they choose to do this, and why 100rpm?
What is the significance of taking the temperature at each site? I assume it is to collect data that is comparable between the different samples, but am unsure what exactly any temperature variation (or lack of) would signify.
Rod-shaped bacteria are significantly more common than other shapes in this study as well in historic studies. This paper provides evidence that it is likely due to the methods used in enrichment and subculturing. They also mention that rod bacteria can divide faster and have better contact with the surface. Does this imply that if the samples were taken from, let’s say, 2-3 inches down in the ground, that the majority of the bacteria would be cocci, spirillium, or other? If different methods were used in enrichment and subculturing, would the rod shaped bacteria still be most prevalent even if the sample were the same?
Figure 4 demonstrates that PAH concentration is reduced significantly after 15 days in incubation. Each chromatogram displays a spike between 15-30 minutes in regards to abundance before dropping back to almost 0 (x10^4). However, there is a slow exponential growth present at the end of each of these figures. Why would this be present?
Each isolate was identified by partial 16S rRMA sequences. Why did they use partial vs. whole sequences to identify? The similarities are mentioned shortly after, would the identifying partial sequences then be the portions that vary by isolate? If there was a 100% similarity between two isolates, should they not look at the whole sequence to make sure there is not genetic variation elsewhere?
Isolate LC is noted to be a human pathogen, and did not grow well in the study. However, the isolate had grown well enough to be sampled in one of the areas surveyed. Why then, was this sample not tested further? Many places where biodegradation is necessary are already hazardous to humans, so the addition of isolate LC, in an area where other isolates are not effective, could be plausible.
Hi Rachel,
I too noticed the reference to naphthalene. I do remember naphthalene was used in the experiments from the last paper we discussed, as a PAH tester. This coincides with the information you found via google.
With our previous research paper we discussed, it looks like hydrocarbon degradation in regards to PAH is under-researched in general. Because of this, I find your questions thought-provoking, especially the one regarding depth and temperature.
I also had difficulty understanding the “killed controls” at first. I believe that they were killing microorgansims, so that any changes can be attributed to the experiment at hand as Professor Ni Chadhain noted. I think they likely do so using one of the methods we recently learned about in our lectures.
Here it notes that only the replicate incubation with the fractions containing high amounts of DNA were used for analysis. I assume that this is to promote a higher chance of favorable data, however, would it change the nature of the data if fraction with lower amounts of DNA were utilized?
I was also intrigued by the method of neighbor-joining. From my understanding (as well as the professor’s comment) the largest advantage of it is the relative quickness as opposed to other methods. This would be interesting to compare to other methods we have used.
I believe that even though the presence of PAH degraders was confirmed, the prevalence of the degraders may not be large enough to show a significant improvement on the amount of oil. I think pursuing a way to amplify their prevalence/productivity could be beneficial.
I believe that the implication is that the research on this topic is still emerging, but I may be mistaken. Either way, the lack of genes characterized leaves area for exploration and a viable possibility for future research.
I googled the phrase “prosthetic constituent” and was unable to find a defintion. However, from context as well as the individual denotations of the words, I believe that it is referencing molybdenum, heme, and nonheme iron as components that together make up (along with the b-type cytochrome) the selenate reductase.
I am also unsure about the exact purpose of analyzing the Se atoms, especially in so many ways. I do believe that this was performed after the degradation however.
I googled this and learned that cosmid clones are plasmids that contain the lambda “cos” sequence. It also appears they are commonly used in cloning, so their usage in this experiment makes sense.
This a question I was thinking myself. I believe that putting the cells directly into the environment would be the most effective way to start the reduction, but not the most cost-efficient. Introducing the organisms themselves would be both efficient and cost-effective.
Because the FNR gene regulates in such a way that requires lack of oxygen, the bacteria is best utilized in locations that present anaerobic conditions. This could be deep sea as in the last experiment, for example.
My question here is: how would one obtain further evidence that the FNR regulates selenate reductase during the transition from aerobic to anaerobic growth? Is this something that would be worth a future study?
I find it very interesting that certain bacteria or species can be individually identified using stain techniques and that those identified bacteria can then be grown on a plate in a lab. The most interesting part is what great new discoveries are in store due to techniques like these.
The first strain of Rhodococcus is capable of degrading several compounds that are harmful to humans and the second strain Rhodococcus sp. is capable of degrading some of these compounds at a higher level. I too wonder what the homological differences between the two strains are and I also wonder, how many other strains are there? What can they degrade? How many strains can be created using the original ones and what will they be capable of degrading?
Its interesting to see the temperature levels of the oven during the Gas Chromatography. I would have thought that a temperature of 300 ºC would kill the bacteria strain.
I was wondering why they measured degradation at different concentrations. Is it in order to get a more accurate percentage and rate of degradation? Could it also be that higher concentrations of Fla allows the bacteria to mutate and become resistant to Rhodococcus sp. faster?
It is interesting to see that Rhodococcus sp. CMGCZ degraded Fla almost entirely, but ILCO is almost completely resistant. I wonder what structural differences between Fla and ILCO cause them to have such drastic resistance differences?
Im interested to know what future compounds can be created with rhodococci and which kinds of pharmaceuticals may already include it. I also researched that it is used in some herbicides which makes it interesting to see that something we have done as students in a lab can turn into something so useful for human life and development.
It would be cool to see the different substrates they could add to enhance degradation and know what specific effects they caused in the Rhodococcus as well as the bacteria it is meant to degrade.
We have seen that PAHs can be presence in both terrestrial and marine environments. Considering there are several ways PAHs can be transferred to humans, why do we not see more cases of PAH effects in humans?
After reading the results section of the paper, it seems that the genes are similar but not exactly the same. This makes me wonder if the genetic changes occurred in order to degrade PAHs more adequately, or if the genes were altered as a result of some other evolutionary adaptation.
I find it fascinating that we use alcohol as hand sanitizer to kill bad bacteria and then we can also use a different form of alcohol to separate cells out to study them without killing them.
I wonder if the nucleotide sequence they found matched others in similar experiments. Also, it is very cool that they could have possibly had a different nucleotide sequence if they used chemicals from a different company or of different purities. Such small scale changes can have such massive effects in an organism.
After reading the results section, it looks like the sequence they found does match some other experiments findings. However, the sequences do not match up as similarly as the biologists here predicted. I wonder what happened that caused their prediction to be so different from what the experiment showed?
Tara, I had the same question. Do our degraders also have an aromatic oxygenase gene, or are there other genes that give provide the PAH degradation ability that our isolates contain?
It seems evident that these species containing genes similar to phnC have very similar evolutionary backgrounds. The differences in amino acid sequences make subtle enzymatic changes but the majority of the gene remains complimentary between species. It seems that the ancestor species expanded environments and its genes evolved making several different species depending on the available substrates in the environment.
I find it fascinating how this paper is so much different than the last paper even though they both are about the same thing. It is interesting to see the differences in how Cycloclasticus degrades PAHs versus Rhodococcus sp. and how two different bacterial species can use different mechanisms for similar activities.
Due to it having a similarity in amino acid sequence that is less than 44% and its cluster falling outside of the main cluster, does this mean that the phnA1 gene uses new proteins different from other bacteria strains to degrade PAHs?
I am interested to know why they were unable to get PCR products for the arsenic oxidase genes, what makes this gene unable to produce authentic PCR products?
It is cool to see that Arsenic is used by this bacteria similarly to that of PAH degradation by Rhodococcus and Cycloclasticus in that, both PAH degradation and use of Arsenic in the electron transport chain are mechanisms essential for survival of the bacteria.
I also wondered why they were stored in the dark. Could it be to prevent the UV rays from mutating the DNA? I would think this would not be the reason though because the bacteria live on rocks baking in the sun all day.
What is the purpose not specifying whether the tubes are shaken or not? Would it not be beneficial to either shake or not shake all of the tubes to ensure all tubes are prepared the same?
Paragraph 7 states that these results suggest that both As oxidation and reduction could be occurring simultaneously. How is this beneficial to the organism? Would it not be wasting energy by oxidizing and reducing the same molecules over and over?
The paragraph says that immediate oxidation/reduction occurred when the samples were in light or dark regimens for each transition. So, compared with the rate of reaction as well, what is causing some of the organisms to stop oxidizing or reducing the Arsenic? Are they finding new sources of energy that aren’t dependent on light/dark that they can use more consistently?
After reading through it again along with the comments, it is very cool that the different organisms are helping one another survive passively, simply by performing their needed metabolic activities.
The passage states that light-incubated biofilms abruptly ended activity at 50ºC. Is this due to processes like the heat breaking enzymatic bonds rendering the enzymes non-functional? I thought that biofilms were very stable and could endure more extreme temperatures than this so could it be some kind of defense the bacteria have that shuts down their activity?
Since anoxygenic photosynthesis is an ancient process, I wonder if the bacteria got their ability to do chemoautotrophy by random mutation influenced by natural selection, horizontal gene transfer, or another way.
[Samples were collected from non-diesel-exposed and diesel fuel-contaminated sites, and bacteria of interest were isolated by a three-step enrichment and screening approach, based on their ability to metabolize phenanthrene]
Later in the paragraph they mentioned that they were able to get three isolates that showed promising potential. How do non-diesel and diesel fuel exposed environments affect the bacterias ability to have a higher phenanthrene degradation ability? It also mentions that diesel fuel contains high concentrations of phenanthrene does that mean that there would be a greater chance of more degrading bacteria present in that sample from diesel contaminated sites?
[Diesel oil, the most commonly used fuel in Antarctica, contains a variety of toxic compounds that persist in contaminated soils, including heavy metals like Cd, Cr and Pb; and polycyclic aromatic hydrocarbons (PAH) like naphthalene, phenanthrene, and fluorene]
As I read this sentence, I wanted to know more on what exactly diesel oil is being used for. I found that people use it for everyday things such as making water, generating power for their heaters and lights, powering vehicles…etc. An enormous amount of energy (diesel combustion) is required to empower transportation due to Antarctica’s remoteness. It really puts into perspective how heavily they rely on the use of diesel making is seem not as easily replaceable with other options at this time. Would PAH- degrading bacteria be a temporary solution to this problem?
[Samples were sealed in sterile tubes and transported on ice to the laboratory and analyzed after 2 months.]
Is there a reason why they waited 2 months to analyze the soil samples? Would the time waited before analyzing have any positive or negative effects on the biological activity of the soil?
I found that in order for biofilm formation to occur there must be these three stages; attachment, growth, and dispersal. What could potentially be a reason for biofilm formation to not occur in this experiment? Does this have any correlation to the soil samples taken?
To add on, it could also have been due to the possibility of other PAHs, being used as additional energy sources, that were present in the mixture of the diesel contaminated soil that contributed to the result of not much of a lag phase and a high growth yield seen in S. xenophagum and R. erythropolis.
P. guineae had a high growth yield when isolated with phenanthrene as the sole carbon source and was found that this strain was able to generate robust biofilm growth in the experiment of phenanthrene stained crystals.
This was not the case when isolated with 0.2 % diesel fuel as the sole carbon source. It had one of the smallest yields. Since there is still phenanthrene present in the diesel fuel, could there be something present in the mixture of the diesel contaminated soil that might be inhibiting the bacterias growth in that environment?
I did some research on this and found that the increase in temperature increases the rate of oil degradation by bacteria. I do believe that this microbial bioremediation approach would be beneficial in other (warmer) environments where there are farms, industrial sites, landfills, and/or onsite sanitation systems.
I wanted to know more on in situ bioremediation and found that this technique limits the spread of contaminates from transporting/ pumping away to other treatment locations while the benefits of ex situ bioremediation include the ability to control variable temperatures, aeration, and nutrient level.
[Vibrio species commonly produce one or more siderophores to acquire iron ]
Siderophores are produced and utilized by bacteria as a result of iron deficiency. Are there other agents similar to siderophores that would help with other depleted nutrients in ocean surface waters such as phosphorus or silica?
[we identify other vibrio species that are natural aerobactin cheaters]
How efficient are these aerobactin cheaters? Since this allows them to conserve energy, would they, in most circumstances, out compete microbes that are able to produce siderophores?
I read that there is such thing as a solid CAS agar media that can also be used to detect siderophore production. How is the liquid assay more favorable to conduct in this experiment?
I believe it has to do with release of aerobactin in the V. fischeri ES114 culture fluids that causes competition within that co-culture. The siderophore competitively exclude V. harveyi growth since it does not possess aerobactin production genes. The siderophores act as a transport system.
I believe so. There was no competition between the two when there was enough iron present, V. fischeri did not have to release aerobactin and V harveyi was able to grow. This is not the case in minimal media. V. harveyi is not able to successfully compete with V. fischeri because it does not have siderophores, giving the advantage to V. fischeri ES114 in low iron environments. I also read that siderophores are able to complexes with other essential elements.
That also intrigues me. I would think that a species would only be able to acquire either or siderophore-producing and siderophore-cheating based on iron availability, not both. Wouldn’t this require a lot of energy? How could they keep up in these environments?
This is an interesting thought. I would assume that ultimately the species that were able to outcompete other species in low iron environments would at some point have to out compete what was once the most favorable such as being able to produce siderophores and siderophore cheating, leading to a new and more efficient way. Seems like an endless cycle.
I thought it was intriguing as well. Also interesting, how a wide variety of microbes must have evolved/ adapted to perform steps of the nitrogen cycle and use it as energy or nutrients.
I had a similar question. I read that even though OMZ’s can reach zero and become ODZ’s (oxygen deficient zones), “substantial suboxic nitrification has been reported in many of the world ocean’s major suboxic zones.”
In the introduction, it was stated that naphthalene is common in water pollution and has a poisonous feature that disrupts mitochondrial respiration. So the question I have regards the bioconversion of the pollutants in the water, did it cause any harm to those working in the clean up?
How is the emulsification activity and high cell surface hydrophobicity related to each other in this sense? Did napthalene emulsify because of its hydrophobic characteristics?
Looking at Paper 1- Fig 4, N7 had the strongest growth out of all of the strains compared to the others. I believe that this particular strain has qualities that naphthalene needs to thrive.
Because only gram negative bacteria were isolated in this sample, is there a chance that if you go to the same area with a different type of pollution, would the results be different?
I agree with Gabriel on this, bioremediation, as opposed to using harmful chemicals, will help with the ecosystem immensely and could potentially aid in future oil spills.
The SDS-PAGE will help break down the protein-protein disulfide bonds and will disrupt the tertiary structure of the protein. This brings the folded proteins down to their linear forms to help determine the sequence of the gene.
If the genes for large and small oxygenase subunits are normally located in pairs, what does that mean for the PhnA1b strand that doesn’t have the small subunit?
Since the PhnA4 gene posses the similarity of chloroplast-type ferredoxins, the isolated bacteria will be able to act as an electron donor to nitrite, glutamate, and sulfite reductase.
The holoenzyme of an aromatic ring provides stability for the structure itself. Once the genes were introduced to E. Coli and expressed, does that mean that the phn gene can encode the holoenzyme by itself?
How is it possible that As(III) is able to achieve anoxygenic photosynthesis in photosynthesic bacteria? Photosynthetic bacteria are supposed to released oxygen.
Hydrogen is a better electron donor as oppose to sulfide. Is this due to fact that sulfide had large error association or because hydrogen is naturally a better electron donor?
I’m a little confused here. What is the purpose of acetate in this experiment? In the previous paragraph it states that acetate does not stimulate As(V) reduction. In this paragraph it states that acetate is an electron donor. Does acetate just mark the presence of anoxygenic photosynthesis or does it do something more?
bioremediation can use microorganisms that are already present or introduced to the environment, does this improve the cycle of pollutants or does it somehow interfere sometimes?
There are many different micro organisms that can be used. Some of these include Pseudomonas putida, Deinococcus radiodurans, Nitrifiers, and denitrifiers.
Narrowing the list down to the best degraders would save time for further examination and begin the process of finding which bacteria would be the absolute best for naphthalene degradation.
To me this sounds like the first experiments labeled certain naphthalene degraders. How did they do this without isolating these bacterias? Since the rest of is paragraph states that this second experiment is the first to isolate naphthalene degraders.
Definitely more harm than good. As a species, we are terrible about exploiting and depleting natural resources with little to no ways of replenishing them. At the rate of the ice melting compounded with more pollutants, we may cause irreversible damage. At point what will we have gathered enough ice to prevent ecological damages? How will we ever conclude were to draw the line?
[Diesel-fuel contains high concentrations of two- and three-ringed PAHs, including phenanthrene. Since it exhibits higher solubility than others PAHs in water ]
It would appear that perhaps the combination of the high concentrations and higher solubility of the diesel-fuel contaminants assists the bacteria as there would be an abundance of resources for them to feed from. As for your second question, I am not so sure myself. Over the summer, I got into brewing alcoholic beverages. In my research, I found out that too much sugar could have negative affects on the yeasts. Granted, yeasts are from another kingdom, but the same biomechanics would apply to bacteria.
Luria-Bertani broth supports a quick and steady growth, however, it is carbon limited. This means the population will plateau quickly, and then the population will decline.
With room temperature being optimal for growth, I would imagine that temperatures warmer than room temp, would cause the bacteria to a bit too quickly, possibly hindering the chemotaxis assay. Some growing colonies may block pathways for other bacteria trying to swim to the chemoattractant substances.
Figure 5 shows how the deletion of iutA or fhuCDB genes causes the two strains’ growth to be completely hindered. What is it about these genes that makes them essential requirements?
From what I gathered in figure 5, it appears that all of the genes are necessary. It’s highly likely that they all work together and missing just one could completely inhibit the strains.
They are more important for the reason of being able to fix nitrogen in an extremely low oxygen environment and they account for more than the “usually less abundant AOB.”
The anoxic layer has more to do with differences in water density. Since both layers never mix, the anoxic layer maintains a low oxygen concentration while the upper layer is free to cycle oxygen.
I believe so. High concentrations of naphthalene are toxic. The data seems to reflect that too little and too much naphthalene affects the growth. I would think they could isolate the ones that thrived above 400ppm and harness their degrading abilities. That way they can have a understanding of the bacteria that can handle pretty much any situation dealing with the degrading of naphthalene.
I am glad that organisms exist that can help degrade oil pollution in the waters. Until this class I did not know that there was such a thing. Is there current research on weather or not other organisms can be introduced to the affected environment to help further degrade the accumulated microbial biomass once the naphthalene is degraded?
It would be interesting if we could introduce the degrading bacteria into the diet of the aquatic life in the affected environments so that there will not only be a constant supply of the degrading bacteria but also so that the other organisms could potentially keep reproducing these bacteria. Is this possible?
I believe the samples need to be shaken throughout the incubation in order to ensure there is no settling of the particles. If they are constantly agitated then there is very low chance that a portion of the sample was not given the same amount of exposure to the NP degrading bacteria. Therefore reducing the chance of the data being skewed.
In order for PCR to work high temperature must be used to eliminate hydrogen bonding between bases and the newly synthesized bases. You need to denature the strand(high temp), anneal the primers (lower temp), and then allow for synthesis to occur and then repeat the cycle.
Since this is the first report on isolation, have these selected bacteria always possessed this ability to degrade naphthalene, or is it something that they were able to quickly develop since it became a rich carbon source that was not always present in these concentrations?
It looks as though BH medium is used to isolate microbial hydrocarbon deterioration. only those able to decompose hydrocarbons will grow. It is designed to add your hydrocarbon of choice for your particular study, which is perfect for this experiment.
Was the antibiotic that was selected something that the E. coli, were resistant to? Therefore making it a substance that the microbe would like to take up and use it for its own benefit?
Chemolithoautotrophs are organisms that obtain their energy from the oxidation of inorganic molecules and it typically aerobic. Photolithoautotrophs use light and an inorganic electron donor. These would be the organisms that could use the toxic form or arsenic for their energy source, correct? I assume are the two types of organisms this paper will be focusing on based on the next four paragraphs.
Are they not focusing on oxic conditions because is has already been manipulated in lab? I think they may be doing so because metabolism under those conditions may be too predictable and not yield a very “interesting” result.
Since their focus is on the anoxic conditions, I think they are trying to see is these organisms are able to give insight on how things were way before there was atmospheric oxygen. I believe arsenic is so toxic to use because we are dependent on oxygen and so maybe this study is trying to reveal how organisms operated before oxygen was even a possible to be included in a metabolic process.
Based on the figures about 45 degrees Celsius is the optimal temperature for maximum As (III) oxidation and As (IV) reduction. This would account for the abundance of the red springs, correct?
Did they decide to disregard the green samples? I do think that the red samples will give out more results to work with since they exist primarily at the optimal temperature for the strain. And I think they have plenty of experimental information from other studies on the cyanobacteria.
I was thinking the same thing. If the DNA responsible for the degradation is in a plasmid that we can isolate then we should be able to genetically modify similar organisms that may not necessarily be able to degrade PAHs. We could then recruit many other organisms and use them to our advantage in this case.
Once the DNA sequence for the degradation of the phenanthrene is obtained could it then be introduced to organisms that can only breakdown NP? So if these organisms were introduced to an environment that contained both, could it then degrade one after it is done degrading the other?
Are there ways to amplify the production of catechols in the cell since the are essential to breaking down PAHs? If so could you introduce different gene sequencing or other non harmful chemicals to the environment to increase this production?
I would like to see if we could mimic these chemical pathways and enzymes out side of the cell so that we can introduce the batch of chemicals to a contaminated area without have to rely on a living organism to help bio remediate contaminated environments.
In the figure the oxidation of As(III) and As(V) reduction are proportionate to each other through out the time of the experiment. Arsenite being the electron donor, and arsenate being the electron acceptor. Arsenate can be used as the terminal electron acceptor. Starting off in the light Arsenite oxidation is favored and in the dark arsenate reduction is favored.
It looks like they wanted to show that using a different carbon source this one being acetate, shows that the organisms do not favor oxidation in the dark at all.
Since these oxidation and reduction reactions are occurring in the same place then there must be a mixture of organisms that are metabolizing these species of arsenic. In the light it is likely that it is an aerobic phototroph and in the dark it is most likely a chemolithotroph.
Could it be that the redox potential for H2 and H2S is higher than that of Ar, that causes the organisms to prefer to use the Ar species as their electron acceptors and donors?
I believe that since the microbes that are reducing in the dark are non photosynthetic then they are not relying on light to carry this out. In the light the temperature is usually in a smaller range than in the dark, this is my best guess.
That is very interesting! I did not know that saline liquids (think fracking) are used to extract hydrocarbons. This is a very important and interesting research topic as many well-studied PAH and BTEX degrading strains do not tolerate high-salinity environments. The high salinity could prevent many strains from growing and ultimately bioremediate and so identifying halophilic and halotolerant organisms is crucial for bioremediation in salty environments. Mannitol salt agar can be used to select for halophilic organisms.
That is an interesting point, Avril. I wonder if we helped efficient bioremediating halophilic strains by adding nutrients etc., if also beneficial compounds would be degraded. For them both are just food sources. Also, some strains may be (opportunistic) pathogens that could potentially harm the environment and the animals living in it. I guess we would have to look at the pros and cons and evaluate.
That is a good question. Maybe start with an enrichment culture which could enhance the density of organisms that could utilize phenol as an energy source for example and then use mannitol salt media to determine if it was halophilic or halotolerant. In this paper, the sample was collected from a salty and oil impacted area so it can be assumed that they can degrade hydrocarbons and are halophilic but it would need to be verified. When it comes to the individual compounds that the Arhodomonas sp. Strain can degrade I think they would have had to test this after the isolate was isolated.
Luria Broth is a nutrient rich medium often used to grow up a variety of bacteria. Minimum salt medium does not contain a lot of nutrients, but we added indole and naphthalene making it a differential and selective media. The conversion of indole to indigo (blue colonies) indicates the presence of monooxygenase and works as an indicator of PAH degradation abilities. Due to the lack of nutrients, only the individuals capable of utilizing naphthalene as an energy source were able to grow up. I am not 100% sure but they may have used both MSM (supplemented with 1 M NaCl) and Luria Broth to see how well the isolates grew in an environment with a higher salt concentration. They knew that they would grow well on Luria Broth but MSM with NaCl would give additional information. Finally, they tested the benzene degrading abilities of individual isolates from both mediums.
I found that they used a GC equipped with a flame ionization detector and a DB-1 capillary column. They also used helium as both the carrier and makeup gas. I think that it is pretty cool that they set up microcosms and used the production of 14CO2 to study the strains’ ability to degrade benzene under various concentrations of NaCl.
I would have liked to know how many bottles containing benzene and NaCl there was and at what concentrations. I only know that it ranges from 1 -2.5 M Na Cl. Also, is 24 micromolar the minimum and 34 micromolar the maximum degraded within 2-3 weeks in any given bottle? We know that the benzene degradation ability was tested from 0-4 M NaCl with no growth at 4, 0 and 0.5 NaCl. The maximum degradation rate was 2 M NaCl which is good to know when evaluating bioremediation strategies in very salty environments. It is interesting that the Seminole strain can degrade benzene and toluene but not ethylbenzene and xylene as these structures are very similar. More details about the degradation pathway are needed to figure this out. The isolate might be lacking one or more necessary enzymes to catalyze certain conversions.
I like that they measured the growth of the Seminole strain in addition to quantifying the benzene degradation. That way they can be pretty confident that they are growing by using benzene as the energy source.
I wish they would write a sentence about how efficient strain Seminole was at degrading phenol. I think it is pretty cool that the degradation results match the genome analysis. Could not grow on Benzoate, GA, and hexadecane since no specific enzymes catalyzing the initial step of these compounds was found. But what about CAT? Were such enzymes found but still no degradation was observed?
The one sample t test looks at two means and determines if they are statistically different from each other. In this case, it was used as a tool for comparative genomics between the Seminole strain and 10 other hydrocarbon degrading halophytes. They found 3 COG categories that were significantly different. This method seems very straight forward and I hope to use it in my research project.
Upper and lower degradation pathways denote 2 different routes that degradation of a compound can take depending on which enzymes that are present. For example, in the upper deg. route of benzene the enzyme epoxide hydrolase adds a water to the epoxide making a dihydrodiol. Further down the pathway the muconaldehyde is converted to muconic acic which is actually a common biomarker for benzene exposure.
It is incredible that we are able to use bioinformatics to predict gene functions based on codon similarities to previously described genes. In this case, after proteomic analysis, which looked at upregulated genes while exposed to toluene, 12 gene products out of the 19 ORF’s predicted to be involved in metabolism of toluene was indeed significantly abundant. It is not ideal that some of these genes were slightly upregulated in the negative control, but they had reasonable explanations for it.
I believe bioremediation definitely could contribute to degrading and removing some of the pollutants you are talking about. Although, it is important to keep in mind that there are hundreds of PAHs and there is not one strain (that I know of) that can degrade all of them. Some bacteria will degrade some PAHs better than others and a mixture of isolates are usually most efficient. That would be an interesting location to do toxicity testing and other experiments. We could for example improve the bioremediation by adding nutrients.
What does an angular dioxygenase attach? From figure 1 I can see that 2 hydroxyl groups are added to carbon 1 and the carbon left of it (1-hydro-1,1a-dihydroxy-9-fluorenone). It is interesting that Arthrobactersp. strain F101 posses the genes to degrade fluorene necessary for all the 3 pathways discussed in this paper. Sphingomonassp. strain LB126 only utilizes pathway number 3 (to the right in figure 1).
It looks like this article describes many useful experiments that are very applicable to our class. However, a lot of the procedures have been conducted and described before and so this paper cites them. For example, plasmid DNA extractions, restriction enzyme digestions, ligations, transformations, sequencing, and agarose gel electrophoresis were carried out using methods described by Sambrook, J., E. F. Fritsch, and T. Maniatis. 1990. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. Thus, to get the methods we would need to follow the citations and find the primary research and publications.
I did not know what primer walking was, so I googled it. It is used to clone a gene from its known closest markers. After sequencing the first piece the following piece is sequenced using a complimentary primer to the end of the first piece. Also, having a primer table instead of writing out the sequences in the previous paragraph may have looked cleaner and better.
It is surprising that no primers could amplify conserved domains of previously described PAH dioxygenases but great that dibenzofuran 4,4a-dioxygenase was identified from a gram-positive bacterium.
I did not know that a transposase suggests that the adjacent gene cluster was acquired by horizontal gene cluster, but I guess that could make sense. orf2 encoded a truncated transposase but no change in GC content was noticed. Definition for a transposase is that these are enzymes that identify the inverse terminal repeat sequences within the DNA and proceed to bind and excise the DNA transposons in between the terminals.
I had to read this paragraph many times to make any sense of it. Table 2 essentially shows the substrate range of FlnA1-FlnA2, from Sphingomonas sp. strain LB126 as expressed in E. coli, and it was compared to 2 well-studied angular dioxygenases abbreviated DFDO and CARDO. When fluorene was the substrate these are the 3 oxidation products: Dihydroxyfluorene, 1-Hydro-1,1a dihydroxy-9- fluorenoned and Fluorenol-dihydrodiol. 1-hydro-1,1a-dihydroxy-9- fluorenone (63%) was the main product and was identical to that of the DFDO mediated oxidation product of fluorene and the CARDO-mediated oxidation product of 9-hydroxyfluorene. However, CARDO does not yield the main angular dioxygenation product when fluorene is used as a substrate which was expected. All the products produced when the substrate was naphthalene and biphenyl also formed by DFDO and CARDO.
After reading this paragraph again after today’s lecture I understand it much better. I am sure it was tough to see flnA1 give a slight band when grown on glucose. I am sure the next phase took quite long as the authors had to repeat multiple steps to find the ideal growth conditions yielding a larger proportion of the FlnA1 and FlnA2 proteins in the soluble fraction. Finally, to confirm the results no oxidation products were detected in the GC-MS when a strain without these genes were grown under the same conditions. This method seems time and labor intensive but very fascinating.
Growing Sphingomonas sp. strain LB126 on other similar compounds is a great way to assess substrate range of this pathway. So, when grown on dibenzofuran it was transformed into 2,2,3- trihydroxybiphenyl by FlnA1-FlnA2. Is this the first step of was it first transformed into the unstable hemiacetal? Angular oxidation products were not detected on either carbazole and dibenzothiophene. The activity of A1-FlnA2 may have been too low. Another referenced paper also found that their isolate could not perform angular dioxygenation of carbazole either.
It is always interesting to research something has not been studied much before like PAH degradation by marine bacteria. I wonder if there is a specific reason for isolating a bacterium from deep sea sediment from the Indian ocean. Had the area been affected by an oil spill maybe. We are probably going to recognize some of these methods from the first two papers.
I could not remember what non-alternant meant so I googled it and it is π-Conjugated carbocycles containing odd-membered rings. High molecular weight PAH is generally more challenging to degrade so I am interested to see how efficient C. indicus can degrade it.
Illumina Solexa sequencing yields short reads up to 100 bp. PATRIC predicts protein coding sequences after annotating the genome. Island Viewer only accepts genomes in GENBANK and EMBL format. Following annotation, we receive a document in embl form. Could we use this on the island viewer website?
I think gene knockout would be better to use if you do not know the function of some novel gene. It is a great confirmation experiment by looking at the difference with and without a gene. However, I feel like RT-PCR is easier and less complicated to do. I would feel more confident that I have found the gene of a certain function if I did a successful knockout experiment. Partially because previous papers have mentioned various reasons for why a gene appears upregulated when not expected to. Leftover mRNA etc.
G+C content can be used to identify regions of horizontal gene transfer. The G+C content can vary from 25 to 75%. The complete genome of C. indicus P73T is 65 mol% and contained 4827 predicted protein-coding sequences (CDSs). It is odd that no gene for tRNA-Tyr was found when the corresponding tyrosyl-tRNA synthetase gene (P73_1712) was identified.
Figure 1 displays the Circular maps of the chromosome and five plasmids of Celeribacter indicus P73T. I like the circle for the chromosome but the circles representing the plasmids are so small that they are difficult to interpret.
It is very interesting that 37 GIs were identified by IslandViewer and it definitely makes me want use it for our project. Can PATRIC identify horizontally transferred genes (HTG) in our bacteria’s genomes? It is interesting that non-essential genes such as PAH degradation genes were acquired through HTG and is likely helping strain P73T’s ability to degrade PAHs.
The plasmids do not contain a lot of DNA. All five plasmids together make up about 9% of the total CDSs in the genome. The arrangement of four COG functional groups were quite different in each plasmid indicating multiple origins and a specialized function for each (PAH degradation).
rep and parA genes were used to do a phylogenetic analysis. I did not know what parA was, so I googled it. It is responsible for plasmid partition. It ensures the proper distribution of newly replicated plasmids to daughter cells during cell division.
I am surprised by how many ring hydroxylating dioxygenases the genome contains. These proteins are multicomponent bacterial enzymes that catalyze the first step in the oxidative degradation of PAHs. Is having multiple genes code for the same enzyme going to make the bacteria better at initiating PAH degradation?
It is clear that strain P73T have acquired genes through lateral gene transfer as region B of the genome is absent in another bacterium of Celeribacter strain B30.
Despite the number of PAH degrading genes found, only the C-7,8 dioxygenation pathway involving extradiol cleavage of 7,8-dihydroxyfluoranthene was used. Thus, strain P73T can be useful to study this pathway in particular. I think extradiol cleavage enzymes incorporates one O2 next to one of the hydroxyl groups (not in between) and activates molecular oxygen.
Since bioremediation is the best restorative means to combat PAHs in ecosystems, does it permanently restore those ecosystems back to their full potential or does it temporarily revive them? Also in regards to Fluoranthene being carcinogenic, what disadvantages are there to using it?
What are other supplemental inorganic nutrients that enhance PAH-degrading microorganisms? Also does having low amounts of YE and glucose limit the degradation potential?
Tim,
I was wondering the same thing initially. However, now that we are writing our summary I am under the assumption that these nutrients don’t always enhance PAH degradation compared to it’s normal ability.
What is the main difference in using two different enriched culture ratios in YMSM? Why was the 1:50 ratio chosen to maintain the enriched culture every 16 days instead of the 1:10?
Why was the degradation such a huge jump between the Fla and the Nap/Phe PAHs? I would have assumed there to be a difference in degradation between Fla and other PAHs but not this significant.
It’s interesting to me that there is no degradation delay in the YMSM medium while the MSM medium delayed degradation by 24 hours. I wonder if the pH, cellular transport properties, and/or chemical structure play a role in the rate of degradation?
It is evident that clear zones forming on MSM agar plates is a technique used for identifying hydrocarbon degraders. But is this technique the best suited for screening or are there other methods that show better identification?
It’s unique that rhodococci can be applied in environmental remediation, in pharmaceutical, and chemical plants. But what is the overall sufficiency of any PAH degrader and is it worth utilizing?
Since Cycloclasticus contains a variety of aromatic hydrocarbons do we most commonly or only introduce this bacteria to marine environments today? Is this the most productive biodegrading bacterium?
I wonder how often there are found cases where a person has experienced life-threatening symptoms by consumption of seafood contaminated by PAHs? Is there a method for seafood-distributors to screen for PAH contaminants prior to market?
Aubrianna,
Honestly this has been resonating with me from the beginning as well. You hear precautions on just about everything else through the news and commercials. Though I have never heard anything regarding PAHs.
Even after completion of reading both papers 1 and 2, I am still not certain which strain of bacteria would have a more beneficial degradation ability.
What is the purpose of noting where the chemicals were manufactured? Is it necessary for other researchers to use these exact companies to replicate this experiments results?
I was thinking the same thing when I first read about their use of the lac promoter. I also was trying to visualize how they would use lac operon’s activating and repressing abilities to help them with their study.
I find it interesting that the amino acid sequence of PHnA1 showed a 51 to 62% identity match with the sequences of PAH dioxygenases while other subunits showed a much a lower identity match. What would be the major factor of this conclusion?
Will PAH degradation decrease since there was no small-subunit gene found in the flanking region of phnA1b and typically oxygenase subunits are in pairs?
Jesse, that’s a great observation! I never thought to think that varied gene location in these species could relate to different habitats. This could strongly correlate that one may have a greater PAH degrading ability over the other.
Lameace,
I too agree that this seems like a much different result than our first paper. PhnC seems like it would be a much greater degrader of PAHs both LMW and HMW.
Lameace,
I too think that this is interesting considering we know the extreme toxicity that arsenic, arsenate, and aresenite bring to the environments. I am interested to know how can bacteria utilize those oxyanions that are so toxic to create more energy without dying off?
Since the researchers were not able to retrieve viable PCR products for aresenite oxidase genes could that mean that the wrong PCR conditions were used or possibly the DNA template has degraded?
Hang,
I agree that the environments characteristics can lead to vital information for researchers to discover certain metabolic reactions. I also researched more on other reasons that scientists believe the habitats are of importance. A new idea is that specific environments are playing a major role in antibiotic resistance globally.
I think it is strange that the temperature ranges were very different. How could the dark As(V) reduction occur over such a wide temperature range while light As(III) oxidation happened over such a limited range? I would think that they would have more resemblance in their temperature ranges.
How could researchers further test to confirm or disprove that the result correlated with the inefficacy of the primers or due to an aoxB-independent mechanism?
Jessica, I too was wondering why the temperature range wouldn’t be consistent between the two. Also, I am curious to know what other environmental changes and/or metabolic changes could be responsible for driving light and dark oxidations/reductions?
Jennifer, I was wondering the same thing. Is it an environmental factor or a metabolic function that is causing acetate to have no known effect on As(V)?
When measuring the biodegradation ability of the naphthalene degrading bacteria, is the speed of the process or the purity of the end product more sought after?
It’s always great to see research that is trying to make the world a more ecologically self place. Future research in these areas will help with any future oil spills.
The fact that only Gram negative bacteria were found is interesting. What would the environment look like for it to have only been Gram positive bacteria? Is that even possible?
A dendrogram is “a tree diagram used to illustrate the arrangement of the clusters produced by hierarchial clustering”, according to Wikipedia. They are commonly used to illustrate the clustering of genes.
It’s interesting to note that the researchers did not find a significant function for orf7 and no obvious homolog and the fact that it is the only ORF being read from right to left. The rest of the found ORFs are read from left to right.
What determines whether the uncontaminated soils from going through As(V) reduction or As(III) oxidation upon arsenic oxyanion amendment? How it uses arsenate as an electron accptor or arsenite as an electron donor?
A halophile, according to els.net, is a “salt-loving organism that flourishes in saline environments”. The Archaea found in the 16S rRNA gene clone libraries are all related to extreme halophiles and are likely present due to the high salt content of the Paoha hot spring environment, as the paper states.
It would be interesting to see any adaptations in electron donors from the biofilms if the hot springs were to ever change or become polluted to the point of mutations being necessary to survive.
So this is one reason why there is no life on Antarctica because of these pollutants. Earth is in danger because these pollutants could are increasing global warming.
Using microorganisms sound like an effective way to help keep the soil of Antarctica in good condition. Wonder if there were different approaches before the idea of using bioremediation.
The ex situ process seems like a better, but longer and time consuming process. The in situ process seems like an ineffective process. Are there any major differences between the two when deployed and which is being utilized most often?
What other group of bacterial strains from these communities have been found to aid in bioremediation? How many hydrocarbon degrading bacteria have been found so far?
What was the purpose of taking samples from non diesel-exposed control sites? Perhaps to gather a statistic of the different phenanthrene degrading bacteria in those areas to the diesel-exposed sites.
So biofilm was used as an indicator to stain the cells of the phenanthrene degrading bacteria? A scanning electron microscope had to been utilized to view the cells’ surfaces. Was this process a necessity since a phenanthrene adhesion assay was done?
I wonder what the effects of bioremediation would have been like if foreign organisms were allowed and if the process would had been harder and longer to accomplish.
I wonder if scientists could ever balance the recurrence of human activity whose to blame for these PAHs and keeping the numbers of these pollutants low.
So caesium chloride gradients help the DNA remain undamaged and allow DNA densities of 3.99 g/cm^3 to migrate towards the top. Ethidium bromide is the dye that makes DNA appear as fluorescent bands.
My question is if these bacteria are able to help reduce the pollutants produced by diesel fuels would we be able to use the bacteria in other places in the world where the population is denser and an extremely large population is? Also would these bacteria be able to survive in other ecosystems or would they have to be adapted for specific regions of the world?
I agree that for years and years mankind has been causing harm to the planet but I believe this was due to the ignorance that we had about the science of the world. Although humans are the most advanced species on the planet we don’t have the answers to everything but as we find things out I think we do try to make changes in our shortcomings. For instance, in our advances in electric vehicles rather than continuing gas powered cars.
How would they ensure that the bacteria were not exposed to any diesel in the control sites. I assume that since the antarctic is extremely cold it probably would not be a very viable option to travel without some sort of transportation and also if they did manage to travel without a vehicle since Antartica is fairly desolate wouldn’t all of it have the same exposure to the diesel?
I imagine it just like with any other living things. Certain requirements have to be met for that thing in order for it to grow. For instance some plants can only grow at high altitudes and some at low altitudes everything has a sweet spot.
I find it very satisfying that this strain of bacteria shows promise in the fact that it can potentially help clean up the mess we’ve made with the use of fuels like diesel. With the help of these microscopic organisms we can make our home clean again.
I understand that these bacteria are specialized for cold weather regions since they were found in Antartica. But would you be able to manipulate these bacteria to continue working in all climates? I think the possibilities for these bacteria could be much larger than just cold weather climates.
Yeah for such a desolate place it is amazing to know that there are still so many different kinds of life even though they are so small. I wonder if finding life in such a cold place could alter what qualifications a planet should have in order to find life.
How do they study these samples of bacteria? Meaning since they come from the deep sea and are adapted to these high pressures as well as temperature how do they transport and gather these samples?
The exploration and understanding of new microbial oil degraders could help us find one that is more efficient at degrading oil and could help with clean up of major spills as to not cause a major impact on any ecosystems.
I was wonder do micro organisms do well with extreme changes in pressure? For instance in this study it says they took the samples and immediately sent up.
I see that in this paragraph they seem to be doing what we were doing in lab a lot with basically trying to get samples from cultures that had positive results. They took these samples and stored them much like we did but why did they end up freezing them at -80 C was this also part of the test or was this just a way to store them for use later?
“The vent fluids, which are laden with petrochemicals, migrate upward to the sediment surface, thus providing a natural model system for studying the microbiology of deep-sea hydrocarbon-degrading communities.”
This is a portion they stated in the introduction describing why they chose this area. Not necessarily due to frequent ships but because of the naturally occurring communities growing there.
I am sure there are other bacteria out there that would hold similar capabilities as Cycloclasticus. Especially due to the fact that there is still so much of the ocean that has not been explored. On top of that micro organisms are not visible to the naked eye so knowing where to look for these things just add to the complications of locating similar organisms.
I agree things that humans see as toxic there could be some organism that thrives on that substance. The fact that micro-organisms are so abundant there are most likely thousands of organisms that we have not even discovered that could survive on alien planets.
Does the red pigment these bacteria produce indicate that they use this method of chemoautotrophy rather than photosynthesis just like most organisms that perform photosynthesis are green?
“The resulting slurry was collected in 150-ml serum bottles as thick suspensions, which were bubbled with N2 and crimp sealed.”
I was was wondering about the statement above. What do they mean when they say it was bottled as thick suspensions and bubbled with N2? Is this a method of storing the bacteria?
I don’t see anywhere that it says the mixture was higher or lower than the 9.3 pH. I believe what they were saying is that they were trying to achieve the same composition of the hot springs so in order to achieve this they used HCl to get it to the same ph. It didn’t make it more basic.
how do scientists know that something like anoxygenetic photosynthesis is an ancient process? Does this process leave some sort of impression or something of that nature?
The large errors associated with the sulfide-amended samples were likely due to the variable kinetics of formation and destruction of various thioarsenic intermediates which are soluble at this high pH (7, 20).
What does this statement above mean? What are variable kinetics that would affect the experiment?
unfortunately i read the end before the beginning..but had i read it accordingly, id have the same question as the others? this experiment seems to serve a minute purpose. we all want to know whats the outcome?
My first question is why did the samples need two months to be analyzed? Then next question is, only two samples? they’re very specific about the cleaning of the spatulas. is there another cleaning method that would disturb or perturb the results? In the Antarctic, it’s fairly cold, so now the question is raised, is there are bacteria that is specific to the area that would interact with diesel a certain way?
Is the soil in Japan deprived from vast amount of Oxygen? because the oxidase product is not isolated here, so does that mean the gene is not converted or is it because of the vast amount of arsenic in the area.?
im in agreement with Autumn here. I like the idea of balance here for the chemical version on respiration or photosynthesis. I am curious of which of the two states are more significant in this biogeochemical cycle.
if the primers are not perfect matches, does that not point to a mutation at some point? Because there should be a level of success otherwise. Mutated strands usually do not have the same capacity as its original.
i find it odd the color correlations here. Green colored water makes me think of swamps. I have been to the west coast and have seen the green waters there and they don’t seem to be a high temp. So the question here is, are the bacterias this color because this is the temperature they become dormant at? like is this color a resting stage for them?
“Recent genetic analyses of the aromatic degradation pathways in PAH-degrading sphingomonads have increasingly revealed that most of the genes necessary for degrading an aromatic compound are scattered in several clusters and not organized in coordinately regulated operons”
The results of the last paper demonstrated similarities in the catabolic gene arrangement of sphingomonas sp. strain LB126 and Terrabacter sp. strain DBF63, the latter a gram-positive strain. In general, do gram-positive strains display a more coordinated arrangement of catabolic genes than gram-negative strains? Are these characteristics generally expected/revealed when studying various strains?
“Nevertheless, the biodegradation of the compound has been poorly studied, especially in terms of bacterial catabolic genes.”
Why is this the case? It would seem that the abundance of these molecules and their carcinogenic properties would merit more interest in understanding their degradation. Is this just a relatively new area of study in molecular biology? It would not seem so.
A general question but I am genuinely curious as it would seem bioremediation studies are rather popular.
“The DNA regions flanking the mini-Tn 5 insertion sites in mutants of strain A4 were cloned by digesting total DNA from the mutants with EcoRI, ligating them to EcoRI-digested pUC19, and transforming E. coli DH5 α.”
Why did they clone these particular regions in the mutants?
Regarding the RT-PCR results, am I to understand that they discovered what appears to an operon involved in PAH degradation but due to its inefficiency its questionable whether or not it would be worthy to investigate further? What would be a reason to further investigate the transcriptional units of such inefficient regions?
[The difference in the fold-increase of the mRNA level in the presence of acenaphthene between arhA3 (2.4-fold) and arhA1 (6.2-fold) remains to be explained, but it is possible that another, more inducible promoter(s) exists between these genes.]
Is there something else that would lead to this difference? Aside from the mentioned difference in stability of transcribed mRNA fragments.
[However, the taxonomic distributions of the five plasmid proteomes of strain P73Twere different from that of the chromosome, suggesting the chromosome and the plasmids may have had potentially different origins.]
Is “potentially different origins” eluding to horizontal gene transfer. If not, could this statement still be indication of horizontal gene transfer?
[Notably, region B of the genome, which contained the PAH-degrading genes and were absent in another bacterium of Celeribacter, strain B30, was predicted to have been acquired via lateral gene transfer.]
“and were absent in another bacterium of Celeribacter”
What is this telling us? In context it seems to indicate horizontal gene transfer but I’m not sure how this absence in another strain would server as evidence.
“These observations suggested that the genes involved in the initial oxygenation of acenaphthene did not function in strains AG2-45, AG2-48 and AG3-15.”
Is this implying that the genes did function in the AG3-69 strain? It would seem they did not according to prior sentences. I’m not sure I understand their conclusion here and how it was obtained.
[We postulated that ORF10 and ORF12 encoded an intrinsic ferredoxin and ferredoxin reductase for ArhA1A2 and tentatively designated them arhA3 and arhA4, respectively.]
Is this statement supported solely by the shared homology with the ThnA3 and ThnA4 genes in strain TFA? Is there support for this statement beyond this shared homology?
In the grand scheme of things, is the intention of molecular biologists to develop a strain capable of degrading PAHs in the ocean on a commercial level? This would appear to be the case but how practical or logical is this considering what is currently known about PAH degradation?
I’d be interested in a comparison of results from similar experiments dealing with bacteria species extracted from deep sea sediment of various oceans/regions.
Based on their description of the degradation pathway of fluoranthene in paragraph 33 I would assume that the experiment did reveal genes for PAH degradation. At the least certain genes were assumed to be involved in PAH degradation based on prior research and homology.
[The organization of gene clusters involved in PAHs catabolism of strains P73T and SL003B-26A1 were almost identical, with only a minor gene rearrangement]
Could this be viewed as evidence for horizontal gene transfer?
The use of indole as a color indicator to physically identify a certain strain of bacteria is fascinating. I have worked with agar plates that contains a specific antibiotic to sort out strains of bacteria; however, I never knew about the reactions of the indole and the dioxygenase enzymes turning indigo as a way of identification.
The isolation of Rhodococcus for the degradation of organic materials is very useful. Though it is great for environmental clean up of crude oil, what is the possibility of these bacteria strains causing harm to the surrounding living organisms? Is there a process to retrieve or eliminate them once the organic materials are depleted?
The use of the mineral salt medium is very common for microbial growth culture; however, why use salts? Salts cause a concentration gradient across the bacterial cellular wall and cause water from the bacteria to be drawn to create a balance; thus, shrinking the cells. Although there are bacteria that can tolerate the concentration, why is MSM widely used for most growth culture?
Incubating bacteria at 30C at 180rpm was a new lab technique to me. I was confused on why they needed to be shaken at 180rpm. I found out that if bacteria were left growing still, it would grow in layers, and the bacteria at the bottom layers would be depleted of oxygen and medium. Incubating at 180rpm would ensure that all the bacteria would growth evenly and efficiently to obtain maximum aeration. Some of you might already knew that, but I wanted to share something new that I learned.
Using PCR amplification as a mean for comparing nucleotide sequences and obtaining >80% identity with the databases is interesting. I did some research and the Rieske (2Fe-2S) protein sub-unit has a function to aid during electron transfer within the cytochrome b6f complex. This seems like a very important function and shows that the relevant matches in the database and the Rieske sequences within Rhodococcus have similar electron transfer proteins.
Is there any correlation or reasoning as to why Rhodococcus degraded only the aliphatic fraction of ILCO in both of the MSM and YMSM medium? With the incubation period being two weeks, what if the time was increased? Would that make any difference?
The utilization of three and four ring PAHs as a sole carbon and energy source for PAH-degrading bacteria can be very interesting when looking at it from organic point of view. Three and four carbon rings contain high torsional strain within the carbon bonds. Do these high energy bonds cause more reactions with the bacteria?
With the advanced technology of CRISPR-CAS, could it be possible to isolate the Rieske center region and incorporate it into a specific species of bacteria of choice that does not harbor the center originally?
The amino acid sequences of these species of bacteria are found to have a 5% divergence based on Clustal W. Does this indicate that the bacteria could contain the specific plasmid for the production of these amino acids?
This is a great point. It leads me to wonder if the most efficient bacteria from every environment could be bio-synthesized to have a dioxygenase gene from each bacteria for maximum efficiency in degrading PAHs.
I believe that the presence of the dioxygenase genes on the chromosome instead of plasmids suggest that the genes are essential for the growth of the cell. However, if bacteria cells share genetic DNA via conjugation with plasmids, are Cycloclasticus able to transfer these genes to other species?
[ This suggests that the electron transport proteins of the PAH dioxygenase system may be shared with other redox systems, possibly to maximize the catabolic potential while limiting its genetic burden.]
This suggestion leads me to believe that the transport proteins of this PAH dioxygenase system could act in a similar fashion much like a-ketoglutarate does in the way it gets generated in the kreb cycle and transported to another pathway for use.
The anaerobic cycling of arsenic is interesting. Further experimentation of this could be very helpful if it the genes for this degradation could be extracted and biologically transcribed by other species. This could lead to health related services when people are poisoned with arsenic.
This in-depth description of temperatures and appearance in the site description tells me that is could be very important to identify the type of metabolic reactions that could be happening in the environment that the sample came from.
Great observation! Maybe the dark and low temperature reduces the slurry’s growth so that it could be preserved to use for several months? I’m not very sure either.
This leads me to think about if this specific environment prevents organism from growing due to the clash between metabolic reactions and hypersaline environment, or do organisms just chooses not to live here?
Though these biofilms are found on the Paoha Islands, is it possible to introduce them to a new environment with similar characteristics and screen for activity or are these biofilms natively active?
The experimental incubations at similar temperatures as the hot springs suggest that they are adapted to that specific environment; however, it leads me to think that if we were to selectively grow them on different temperatures, it would eventually adapt and change genotypically.
If scientists use bioremediation in a certain area and the microbes end up not working, what would be the effect? Would they just leave the bacteria in the water?
How are sampling sites determined? I noticed they do not give an exact location and the contents of the sediment is tested and discovered after the fact but do the researchers just go to random sediments and test the soil?
As the researchers determine which microbes grow best on different sources, how would they then be released into the areas where bioremediation is needed? I.e. would they just release a mixture of microbes into different areas in order to degrade everything that might be there or would they release only the best microbes to degrade whatever substance is most present?
So they concluded that Cycloclasticus will possibly be able to degrade PAH compounds but were not able to get definite results in this study? I just want to make sure I am understanding correctly. n
Is biomineralization almost like bioremediation in this case? Since too much Se can be toxic and it is left up to bacteria to mineralize it so it doesn’t get to elevated levels, it almost mirrors how bacteria degrade PAHs in my opinion if that is correct.
When they say that only 2 genes have been found in the two bacterial examples, does that mean that only those two bacterial strains are the ones mineralizing the Se in waters?
Since the E. coli S17-1 rapidly reduced the Se(VI) concentrations, how would scientists use this to reduce pollution? would they use the cells or use the organism to put into polluted areas to reduce the amount of Se?
are chlorate and bromate pollutants? Since the bacteria can use the enzyme to reduce those but not the others, would that still make them useful in finding efficient degraders for pollutants?
Are the organisms they use in these studies able to be relocated into areas that need the selenate degraders for the pollutant or is the main focus of this study to find the families of bacteria that can degrade selenate?
This paragraph was discussing the isolates strain of ZX4 and its effectiveness for degrading phenanthrene. Because it talks about potential one used for biremediation of oil-contaminated enviroments could this be used to help clean up oil spills?
If the microorganisms are used to remove the oil spill, how do scientist know how much microorganisms need to be placed in the oil spill to remove the oil? What is the effect on other organisms with using the microorganisms?
Oil usually remains at the surface, however it does sink down, can the microorganisms reach to a certain point to continue eating the oil at different depths? Has there been more research on the effect of the Oil spill in the Persian Gulf to this day?
Using the Gas chromatography was a way to extracts any condensed by evaporation. Is this a final step of in the scientists methods to isolate the naphthalene from the oil?Why would the concentration of naphthalene be incubated for seven days? The gas chromatography will give the scientist a percentage of the naphthalene.
From moving to different flasks and being filter could that possible cause any cross- contamination while trying to isolate the naphthalene? Could there be any other types of growth in the flasks?
What made the “18 isolated stains that show adequate growth rate” grow faster than all the other strains that were from the marine samples? Also in the beginning the article, it tells where each strain has came from and what they correspond with. For example N1 is a algae strain.
What would cause the adding this concentration to dramatic decrease the growth? As the growth decreases, the strain N7 had its maximum growth, what would cause this strain to have its max growth compared to the other stains?
Was using different researchers to obtained concentration used to compare data with the other researchers. As using the multiple researchers, it showed the E24 is the direct relationship between emulsification activity. What is the significance of E24?
How can isolates be helpful in bioremediation of PAH- contaminated sites and oil spills preserve the ecosystem in a natural way? In other words, what are the steps to preserving the ecosystem with knowing this knowledge?
Will the phylogenetic trees be compared with different phylogenetic trees that are base on 16rDNA? How was the phenanthrene-degrading bacteria strain being isolated?
Why will the scientist use meta-cleavage to catalyzed the conversion of catehols? And what would the scientists find with using the meta-cleavage method?
Why was the lowering of C23O activity observed when ZX4 was growing? Was it because it was using the glucose and using the carbon source on the plates as energy?
If these bacteria were found in a hot spring, would most of them be chemolithoauotrophs if there is a lack of sunlight and use carbon dioxide for its source of energy?
In figure 1, it is showing how the samples shift from light to dark, but how come at about 70 hours the As (III) dies, then start having responses around 120 hours?
How come the activity was 5- folds higher than the comparable dark- incubate ones? Could it be because of photosynthesis and the acetate prefer the light.
If the bacteria degrade these harmful compounds, such as the benzene rings, and then continue to be in the water, will the by-products created through degradation have a harmful affect on lifeforms as well? How long does this method have to be used before we can be sure of any side effects of this system?
Not only would environmental conditions impose selection pressure, but that combining with the uptake of different chemical components could potentially cause mutations within the bacteria used for degradation. How can this be monitored to keep a watch on any potentially mutations that could become harmful and decrease water quality?
Assuming that there is a notable mutation rate within the bacteria during incubation period, the PCR data could be skewed and cause a misread of the rRNA as a different bacteria that is not of importance to this experiment. Could satellite markers have been used in conjunction with PCR to spot possible small mutations?
If the time frame of this experiment were adjusted in either direction, would we expect to see a consistent rate of fixation or is there a peak efficiency time frame? At what point does the degradation become ineffective, either over time or during certain concentrations?
Would the growth of the various strains together create a competition over resources? Is it possible that the less productive strains would preform better in the absence of high preforming strains?
What differences in the strains cause this ability to handle different concentrations? Is this the product of mutations in the bacteria? Were these mutations already present or could they have occurred more rapidly though lab exposure?
With the bacteria coming from such a harsh environment, it seems difficult to appropriately replicate the extreame conditions in a lab setting. Will this affect the amount of oxidation/reduction taking place in the sample?
So instead of draining the waste in the environment can they collect them and treat them with microbes and then dump it outside? If those harmful compounds are abundant in the environment, how much amount will affect the mammals?
PAH bioremediation breaks the compound in CO2 and H2O than the level of CO2 will increase in water. Will this change affect the organisms in water, or there are no organisms because of polluted water?
Degrading PAHs using microbes is easy cheap, but will the overcrowding of that microbes at certain area harm the other creatures around them? Even before throwing the waste out, it can be treated with microbes and then let it out.
The phylogeny tree is always useful to understand the bacteria, as we get to learn which is similar to which and to draw the conclusion on the specific matter. To learn PAHs and their pathway of degrading will help us to understand how some plastic and metal cannot be degraded.
The colonies grew on the diluted plate would be light and when grown on fresh media will they grow faster than diluted as more resources are available to them?
So with the help od content of G and C content of DNA we can find the Species and Genere of the microbes? what if the moles are same but have different placing order?
They were not able to obtain PCR product for arsenite oxidase genes, but instead of what they are assuming, they might be using a different gene or enzyme to reach the last step? Or they can even be the totally different process which is yet to be discovered?
As the reduction of As(V) and oxidation of As(III) can happen in both oxic and anoxic condition, they actually donot need CO2 or O2, and they might be the oldest living creature or one of earliest living things on earth?
There must be some sort of limitation on what we can or cannot do to protect Antarctica from human pollution. Guideline must be put in place to ensure that tourist only leave footprints and nothing else. Excessive mining of fossil fuel shouldn’t be allowed to preserve natural resources.
As I’m reading through the first few paragraph of this article, I notice this type of experiment is what I am learning right now. From the type of bacteria to the agar media. Moving on to creating primer to preform PCR. By using primer 27F and 1492R, the purpose was to make a copy of the 16s gene from 27th basepair to 1492nd basepair? In genetic lab, we called them by the name of the gene, which mean the primer F and R are from the same gene.
Is the primer 27F and 1492R from the same gene? From what I understand, PCR is basically making many copy of a segment of a gene. In this case the gene is 16s, is that mean they are making a copy from 27th bp to 1492nd bp?
I think PCR is rather common nowadays. We are actually preparing for a PCR in genetic lab right now. My group is using the gene name unc_27 in C.elegan.
I’m very interested in the human microbiome and all of the roles bacteria play in/ on our bodies. I’m sure the earth’s microbiome plays a huge role as well.
I’m not sure if they have found anything having to do with what you suggested but we are finding out ways in which the human microbiome is beneficial to human health. For example, bacteria that make up the “estrobolome” help women (and maybe men too.. idk) get rid of excess estrogen from their body. I’ve also read about certain microbes possibly protecting against skin cancer. (I’m not sure if that has been proven. It might just be a theory). Your gut microbiome has a good bit to do with your immune system as well.
I would definitely assume that the bacteria adapted to the diesel contaminated soil, especially since other bacteria were killed off by it.. When humans take antibiotics, many have issues with yeast overgrowth that wouldn’t have had the chance to grow otherwise. The decimation of normal gut flora allows other microbes to grow.
The bioaugmentation was very interseting to read. Additionally, with advancing development or straightforwardly embeddings regions with contamination corrupting microscopic organisms, what might be the effect on the encompassing environment if this process was left unattended?
What was the idea behind the extraction repeating three times? Also, did the excitation-emission spectra make the chemometric computation increase or decrease?
I did my research on CARD-FISH and flow cytometry because I wanted to know exactly what their role was. CARD-FISH can be used for phylogenetic staining of microorganisms in many environments, while flow cytometry is used to enable rapid analysis of significant number of cells at single cell wall. How does flow cytometry count cells?
[This process can be achieved by promoting the growth of endogenous metabolizing bacteria in contaminated sites (biostimulation) or by directly seeding contaminated sites with pollutant-degrading bacteria (bioaugmentation). ]
Is the bioaugmentation process similar to the process that was used cleaning the Deepwater Horizon oil spill in 2010? Also, with promoting growth or directly inserting areas with pollutant-degrading bacteria, what would be the impact on the surrounding ecosystem if this process was left uncontrolled?
[Oil contamination can generate detrimental changes in soil properties, including modifications in maximum surface temperature, pH, and carbon and nitrogen levels (Aislabie et al., 2004). ]
I googled the changes in temperature changes in Antartica, and found out that it is experiencing some of the most rapid warming on Earth. In the last 50 years, Antartica’s temperature has increased over 5 degrees Fahrenheit. Is this due to the reshaped microbial structures?
[Environmental bacterial isolates, as well as the control strains Escherichia coli BW25113 and Pseudomonas aeruginosa PAO1, were routinely grown in R2A and M9 minimal media. ]
Why would R2A and M9 minimal media be the choice of mediums for this research? Isn’t there much better mediums for E. coli, such as Luria Bertani broth?
[After 90 min incubation at room temperature, buffer contained in the syringe was recovered, serially diluted, and plated to estimate CFU/mL.]
With the buffer being incubated at room temperature, what would be the effect of adding a bit of heat. Would the 90 minute incubation drop?
[Samples were taken from the surface soil horizon (0–10 cm) from four sites exposed to diesel fuel, and four non diesel-exposed control sites,]
With all the sample taken from a soil horizon spanning between 0-10 cm shows that these samples are not from a particularly high horizon. Soils with higher horizon are exposed to more organic matter, and less human disturbance. Would taking a nutrient richer sample show a better perspective of how diesel fuel is altering the soil’s ecology?
I find it so interesting that a place like Antartica has a great diversity of bacteria. With such harsh climatic conditions, how is this possible? No trees, shrubs, and very few flowering plants have the ability to survive this ecosystem. Wouldn’t bacteria diversity decrease as the climatic severity increases?
[ Leave a comment on paragraph 4 0 Varied processes have been described to enhance the ability of bacteria to metabolize PAHs, including the formation of biofilms,]
It is amazing to learn all the ways, formation of biofilms help bacteria. I know bio films can physically protect bacteria from antibiotics and disinfectants, but this process takes resources and energy. With such harsh conditions in Antarctic regions; do these biofilms have a change in ability to metabolize PAHs compared to the ones in high nutrient ecosystems?
[ Leave a comment on paragraph 8 4 In environments harboring multi-species bacterial communities, siderophores are known to play roles in competition for iron]
Siderophores have both cooperation and competition interactions in their environment. To high of iron concentration can promote algae growth which can block off sun light and throw off normal feeding processes. My question is, why do siderophores have roles in competition? What is the advantage of siderophores sequestering available iron?
I researched that vibrio concentrations in oysters can be 10-fold higher than the surrounding water. Most shells fishes are great sources for iron. I was wondering could there be a correlation with siderophore aerobactin in vibrio and high concentration of vibrio within oysters?
[ The V. fischeri ES114 genome was scanned for siderophore biosynthetic genes using AntiSMASH with relaxed strictness]
I had to google how AntiSMASH worked for scanning siderophore biosynthetic genes, and found out that it allows rapid genome-wide identification, and analysis of secondary metabolite biosynthesis gene clusters in bacteria. With this being such a broad genome scanning, is there a larger chance of mistake?
[Unless otherwise indicated, erythromycin, chloramphenicol, kanamycin, ampicillin, and polymyxin B were added to final concentrations of 5 μg mL−1, 10 μg mL−1, 100 μg mL−1, 100 μg mL−1, and 50 μg mL−1, respectively. ]
I understand that these antibiotics were added to test the different resistance genes within the genome. My only question is, why are they added at such different concentrations? My guess is that the strength is huge factor when comparing the different antibiotics.
[following boiling or filtration through a 10,000 MWCO membrane, suggesting that the inhibitor is a small molecule ]
The molecular weight cutoff (MWCO) is in important term for the membrane filtration, describing the pore size or rejection ability of the membrane. Typical nanofiltration is anywhere between 200 and 2000 MWCO, so a 10000 MWCO is evident to an extremely small molecule.
[ It was curious that growth inhibition of V. harveyi occurred only when culture fluids were obtained from V. fischeri ES114 grown in minimal marine medium but not in rich medium]
This is a very interesting outcome because for most situations the rich medium would yield the better growth in colonies. Does this show evidence of the advantages of siderophores when iron is low in the environment?
[ Public goods producing bacteria are vulnerable to cheaters, bacteria that benefit from using public goods without paying the metabolic cost of producing them]
I understand how these cheaters have a competitive advantage over public good producing bacteria ,but is there a correlation between densities of both in the environment? If the advantage is strong, why is there a need for the cell to keep these genes that have a metabolic cost to make? I was thinking that for there to be cheaters, there has to be public good producing bacteria close by, and in higher densities.
[. Aerobactin production by V. fischeri ES114 could be especially relevant during colonization of its symbiotic host.]
I would imagine aerobactin production is a crucial part in colonizing a new symbiotic host. For reasons such as colonizing in a time of the year or after long draughts of rain where the iron supply is even lower than normal. Also, if the symbiotic host lives in a deeper part of the ocean where iron supplies are naturally lower than the ocean’s surface.
[Nitrogen losses from these oxygen minimum zones (OMZs) are estimated to account for 30–50% of total nitrogen loss from the oceans (9, 10)]
I always believed that the oxygen abundance would be similar in all areas of the sea. After reading about OMZs in the oceans, is there a correlation of water depth and oxygen abundance in the sea?
[A 20- to 40-m-thick suboxic transitional zone, characterized by low oxygen (<5 μM) and undetectable sulfide, persists throughout the basin between the surface oxic layer and the sulfidic anoxic deep water (≥100 m) (12, 13).]
I was wondering, with sea temperatures rising due to global warming, is this possible reason for the thick subtoxic zone in the Black Sea?
[. 15NO2 − production was measured in the same anoxic 15N incubation vials as in the anammox rate measurements, but 15NO2 − was analyzed as N2 after a two-step reduction by acidified sodium iodide and then by copper at 650°C]
Why was copper used in the reduction steps? Aren’t metals like magnesium and iron a more common metal found in the sea?
I did some reading on CARD-FISH to see the differences between in and FISH. CARD-FISH is a specialized form of FISH focusing on aquatic habitats with small, slow growing, and starving bacteria. It allows hybridized cells that are usually below detection limits to be signaled.
These graphs show distribution of inorganic nitrogen, sulfide, light transmission, and reduced manganese at different water depth, but focusing on the suboxic level. I gathered that from graph A, nitrate was at its highest level at about 82 m. In graph B the O2 levels drop significantly when entering the suboxic level dropping lower than 1 micrometer. Graph C shows that MnOx was at its optimum level in the suboxic zone.
This maximum likelihood tree shows likely hood of a phylogeny evolving are evolving from, and from this tree shows many clades that amoA genes were expressed at 80 m.
[Clearly, the production of 15N15N in 15NH4 + incubations was a result of anammox being linked to nitrifiers when no other NO2 − was readily available for anammox]
I thought the 30N2 isotope could be a product of anammox-nitrification coupling. Does denitrification also occur because if not shouldn’t there be a nitrite product in graph a from figure 2?
Are non-sporulating aerobic rhodococci the most common found contributor to biodegradation because they are not giving all their energy to offspring production or is it something else?
Throughout most of the materials and methods I noticed that a multiple number of processes were incubated at 30 degrees Celsius combined with being centrifuged at 180 rpm for a week to two weeks. What was the reason for why they had to wait so long for each experiment before continuing and was there not a faster way for them to do these?
Even though the amplified PCR products were obtained and registered in a database, if the same soil bacteria sample were to be subjected to another experiment would the sequences be changed because bacterial DNA could be altered through conjugation or transduction?
I’ve seen multiple times throughout this research paper that everything that was extracted was analyzed by GC-FID. Was this the only instrument that they were able to use to get the results that they desired or was there another way to get the results but they didn’t have the equipment for it.
I noticed that the one gram soil sample was subcultured at a 1:10 ratio followed by a 1:50 ratio that was used for the 16 day period? Why was the ratio so low for the first initial step and then was stepped up to a 1:50 ratio for the rest of the experiment? What made it so necessary to have to use such a big ratio? to carry out the rest of the experiment?
For this table, when the researchers were determining what to use for the best relevant match, was there a set percent identity (nucleotide) that they were looking specifically for or was it just based on what matches were the highest that they could find?
What was in the medium that was supplemented by YE that made Nap not able to degrade? Was there a certain characteristic that it possessed to inhibit the degradation of NAP?
I was wondering if this bacterial strain screening process is still used today? I’ve tried to look it up on google but, I did not find any type of information correlating to that particular process.
When they were testing the addition of YE in mediums did they try different amounts of YE in each set of mediums or was there a generalized amount of YE that they would use for each Rhodococcus sp. CMGCZ sample.
I find it very interesting that the Cycloclasticus strains are able to to degrade substituted and substituted PAHs. I’m very curious about the extent of how efficiently they can degrade compared to Rhodococcus.
I agree with your statement. With the algae bloom in peak during the summer in the Gulf coast, research for the degradation of toxic chemicals would be helpful in retaining the marine biome.
I noticed that when the beta subunit was compared to PhnA2 it stated that it had a relatively high sequence identity of 52%. Is this considered to be high because the sequence identity is unable to get much higher than that or is it high because its over 50%?
I was thinking the same thing when viewing this figure, and to make matters worse I don’t understand why orf7 is going the opposite way as opposed to all the other open reading frames.
Someone can correct me if I’m wrong, but I’m pretty sure that the purpose of the flanking region is a region that contains the promoter and also may contain enhancers and or protein binding sites.
Madeline, I am too very interested in knowing if further studies will be conducted for the efficiency of other dioxygenase genes. I also wonder that if they are to find other dioxygenase genes if they will be much more efficient than the known genes or if they will not be as good.
Nolan, I am very interested as well to see how this study will further help the understand of the evolution of arsenic from the anoxic conditions compared to oxic conditions.
It is very impressive to see that there are organisms in this world that are able to withstand environments that would kill anything else when exposed to the conditions. Could these Organisms give rise to something that could be very beneficial to survival or could they just end up being a dead end road.
Taylor, I’m wondering the same thing. I have done extensive googling and I have yet to find another gene that codes for aerobic As(III) oxidation, but I also could be looking in the wrong place as well.
Derria, I was wondering the same thing but my best answer for it would be that maybe they experimented with a various amount of temperatures and found that the set temperature that they kept them at is what worked best for them.
Spencer, I agree that you would expect to see more archaea in this type of environment, and I also agree with you that for once while reading a paper like this that I actually feel as if I have an understanding of what is going on.
Sarah, I was thinking that the reaction was able to proceed at a much faster rate because it was done within aerobic conditions, because correct me if I’m wrong but I thought that anaerobic respiration was much quicker due to the lack of excessive ATP that is produced, but I guess that it would only be quicker if it was done within anaerobic conditions.
Julie, I agree with you, when i first saw the figures in the results section it did not make much sense to me but when I read what was being discussed it helped a lot with the understanding of what was going on.
Although the location had insufficient nutrients and high hydrocarbon pollution, what can be done today to attempt to isolate bacteria from the initial pollution site?
The figure helps to visualize and makes more sense of what is going on in the experiment. As well as show why some isolates grew well on some agar plates than others.
In this step the potential strain were mineralized and grown separately in a liquid medium. Following this the cell’s biomass was washed several times prior to use to ensure sterilization.
In this figure, it shows for pECL32 the transcriptional direction is to the left, gene location to be in fnr and ogt and that it is positive for selenate reductase activity.
I was curious about the Antarctic treaty that apparently prevents “the introduction of foreign organisms into the Antarctic continent,” so I looked up the details. The treaty was set in place in 1961 and holds Antarctic aside as a scientific preserve and allowing freedom for scientific investigation on its land. It does not allow for any military presence or any kind of weaponry testing anywhere on the continent. The treaty allows for Antarctica to be the one place on Earth that is uninfluenced and untouched by human interaction. It is essentially a natural and pure laboratory for the world to use carefully.
I was surprised to learn that they heavily relied on diesel oil in Antarctica as well. It seems like microbial bioremediation would be a good solution temporarily, but overtime I’m afraid with the increasing issue of global climate change that it would not be enough to keep up with the growing contamination and pollution. Especially as human activity only continues to grow. I am under the impression only so much can be done with the limitations Antarctica is under as the article states “bioaugmentation can only be implemented by the use of native microbes.”
I was curious about the other methods use for quantifying chemotaxis other than the modified capillary assay mentioned in the article.I found that other methods use bacterial concentrations that are too high to inhibit chemoeffector consumption or require conditions that make quantifying too burdensome. The technique of modified capillary assay for quantifying chemotaxis is actually an inexpensive procedure and can be apparently used with volatile or semivolatile chemicals.
Non-polar compounds were chosen to extract from the culture media. Extraction was repeated three times. I wondered why in this particular step of phenanthrene quantification, only non-polar compounds were extracted instead of polar as well and why specifically three times for extraction?
S. xenophagum D43FB and P. guineae E43FB were both shown to have the ability to adhere to phenanthrene crystals, however only D43FB was chosen to further study by scanning electron microscopy. Is there any particular reason why this isolate was chosen over E43FB even though they were both shown to exhibit strong and intermediate staining?
[Finally, although D43FB failed to show chemotactic responses, several genes encoding components of the flagellar system were found, suggesting that this strain has the potential to swim and respond to attractants in its environments, under the proper gene inducing conditions. ]
It is apparent that D43FB has strong potential as a good PAH-degrading bacteria candidate for a bioremediation, I however am not fully convinced it is truly the best candidate. The next question or set of experimentation for the researchers is to see if the strain can perform well under the conditions found in Antartica. My concern is that it will not perform as expected when the carefully crafted conditions created by the laboratory dissipate when they are tested in all actuality.
[While S. xenophagum D43FB was unable to produce biosurfactants and did not exhibit chemotactic responses, this strain exhibited the ability to form robust biofilms in vitro and was able to adhere directly to phenanthrene crystals, as shown by SEM microscopy, suggesting this bacterial isolate can tightly interact with phenanthrene when using it as its main carbon source.]
As previously mentioned in the article, a bias was found in the experiment because the strains were grown under standard laboratory conditions. While it is great the researchers found D43FB was able to form bioflims, it also occurred in vitro meaning an experiment performed in a test tube or laboratory dish. I would infer that this does not guarantee that S. xenophagum will always possess this ability especially when not completed inside of laboratory conditions, but in the actual environment.
Like many of my other classmates, I too was curious about the different methods of bioremediation. I found one article that lists the advantages and disadvantages of using In situ to ex-situ bioremediation. In situ is typically less costly because equipment is not required to unearth the contaminated soil and it is cleaner overall because it does not circulate dust and pollutants back into the surrounding area. One of the disadvantages is the extended length of time for decontamination. The three main strategies that fall under In situ are Biostimulation, Bioaugmentation, and Bioattenuation.
[The remarkable finding that atmospherically-transported Saharan dust enables proliferation of vibrio bacteria by delivering dissolved iron to surface marine environments further demonstrates the exquisite scarcity of iron in the ocean ]
I too was confused on whether iron benefits or impedes bacteria, and thought looking up the interesting finding of the Saharan dust delivery on the ocean surface would help clear up some confusion. The article I found had a team travel to the Florida and Barbados to assess the levels of Vibrio growth in ocean surface water during natural Saharan dust events. They found in just 24 hours of exposure, Vibrio background level went from 1 percent to almost 20 percent. They also say vibrio has the ability to quickly respond to nutrient plumes at a faster rate than microalgae which could reveal that Vibrio could play a major role in being intermediaries in the biogeochemical cycling of iron.
[Surprisingly, in our initial attempt to characterize the co-culture system, we found that V. fischeri produces and releases an inhibitor that prevents the growth of V. harveyi.]
In the previous paragraph, the researchers said that they chose Vibrio harveyi and Vibrio fischeri because they are known to occur together in nature. My question is why then does the problem arise with V. fischeri releasing an inhibitor and preventing V. harveyi from growing under standard laboratory conditions?
[V. fischeri ES114 makes almost no bioluminescence under laboratory conditions so the presence of any residual cells did not contribute to the bioluminescence reading.]
I thought this was interesting and wanted to know why exactly it would make almost no bioluminescence under laboratory conditions? I researched what could be the cause and found that when in low density like open marine sea, the luminescent genes are turned off.
[Plasmids were transformed into E. coli by electroporation using a Bio-Rad Micro Pulser. ]
I was curious about the process of electroporation so I decided to research it and found that it is the physical transfection method that uses an electrical pulse to create temporary pores in cell membranes in order to increase permeability of the cell membrane. This allows chemicals, drugs, and electrode arrays to be introduced into the cell. Interestingly, it can also be used in tumor treatment and gene therapy since it does a great job of introducing foreign genes into tissue culture cells.
[Thus, the difference in siderophore production between V. fischeri ES114 and V. fischeri MJ11 does not stem from differences in transcriptional regulation, rather, the difference apparently arises at the protein level, perhaps due to differences in post-transcriptional regulation, biosynthetic enzymatic activity, or protein stability.]
I understand that the researchers tested the efficiency of biosynthetic enzymes in V. fischeri ES114 versus in V. fischeri MJ11 by overexpressing the iucABCD operon from each strain in a V. fischeri ES114 ΔiucABCD mutant. The exact cause of this variety in siderophore production is not exactly specified by this test. I was wondering if they were other tests that the researchers could run to determine between all of the possibilities, which one is the determinant?
[qRT-PCR confirmed that luxT was transcribed from the overexpression vector in E. coli as there was a 40-fold increase in luxT transcript levels in the strain supplied with the arabinose inducer compared to the isogenic uninduced strain ]
I was unsure what the exact meaning of “isogenic” was and found that it refers to organisms having the same or closely related genotypes. Concerning the rest of this paragraph, I see that it was essential for the researchers to perform some ground work by testing the limits of LuxT and how it will contribute in comparing and contrasting vibrios species competition.
[First, possession of AerE makes an aerobactin cheater immune to cytoplasmic aerobactin toxicity. We say this based on our assessment of the growth defect displayed by the V. fischeri ES114 ΔaerE mutant (Fig. S10A). Second, possession of AerE may enable aerobactin recycling by cheater vibrios, fostering higher overall iron acquisition and, in turn, a superior growth advantage during competitive situations.]
Just because a growth defect was observed with the V. fischeri ES114 ΔaerE mutant during the experiment, is it fair to conclude that any and all possession of AerE will make an aerobactin cheater immune to cytoplasmic aerobactin toxicity?
[Moreover, both vibrio species produce a variety of public goods including extracellular proteases, chitinases, and QS autoinducers, all of which can be monitored in real time.]
I was curious as to what exactly a”public good” meant exactly in this context. I did a google search and found an article defining it as “biological public goods are broadly shared within an ecosystem and readily available. They appear to be widespread and may have played important roles in the history of life on Earth.” They are essential factors that have played an important role in the evolution of life like merging of genomes, protein domains, etc.
[Nevertheless, the identity and abundance of the responsible nitrifiers, or any coupling between nitrification and nitrogen losses, remain poorly documented.]
Are these couplings poorly documented because it is difficult to record data at such deep depths of the ocean?
I wanted to know more about Crenarchaeota so I looked it up and found that it is known for having microbial species with the highest growth temperatures of any organisms. This is why they have been isolated from and able to sustain in deep-sea marine environments.
[Anammox bacteria was verified by CARD-FISH with the probe BS820 (15), but strong background fluorescence precluded accurate enumeration and qPCR was used for quantification. Total microbial abundance was measured by flow cytometry (57).]
I was curious to see what exactly was flow cytometry and found that it is a technique used to detect and measure the physical and chemical characteristics of cells. Is this method a default when the population of cells are hard to observe?
The CTD system detects and measures the temperature, pressure, and conductivity of seawater. These factors all affect what kind of microbes are present in the various depths of the ocean. I researched what were the main differences between the two types and found that pumpcast enables high resolution water sampling along vertical profiles, while rosette cannot sufficiently detect small vertical structures in the vertical distribution of trace metals.
These various graphs for figure 1 confirm a couple of things. In the introduction the researchers state that the suboxic zone can be characterized by low oxygen and unobservable sulfide. The anammox bacteria levels are high at this zone which also confirms the notion that nitrogen loss via amammox occurs here. Figure 1B and 1D both verify these statements respectively.
These graphs correlate to the figure 1 graphs regarding the nitrification and anammox levels in their respective zones. In figure 2A, it can be seen that mRNA grows exponentially well in the suboxic zone with the ammonia-oxidizing crenarchaea. In figure 2B, we see poor growth from mRNA and BAOB cells. In figure 2C, mRNA did not follow a strict growth pattern, varying quite a bit. YAOB cells grew the most in the suboxic zones.
[These observations may imply that in such suboxic settings, these crenarchaea were not using their nitrifying capabilities much but some other energy-acquisition pathways.]
What might be some of the specific reasons that ammonia-oxidizing crenarchaea were not able to utilize their nitrifying capabilities in the suboxic zone?
[Despite the barely detectable gene abundance, strong amoA expression by AOB was detected within the nitrification zone (Fig. 2). γAOB amoA expression, in particular, was up to nearly 3-fold greater than that of crenarchaea. ]
I find it interesting how the gene abundance of amoA was so low, it still however expresses so strongly in the nitrification zones. Although it was strongly expressed, is it possible that the low gene abundance is the reason why it did not measure up to its predicted net nitrification rate previously set by the researchers?
I find it interesting that the present species Rhodococcus sp. CMGCZ is able to degrade higher concentrations of Fla than the previous species of Rhodococcus. I’m wondering what caused the change between the homology of each species and what changes to the homology makes the present species different from previous species?
Since the present species of Rhodococcus is able to degrade high concentrations of Fla would that change the cycling of the pollutants or interfere with the function of marine and terrestrial ecosystems?
When I first read this paragraph the first thing that caught my eye was bead-beating, and so I decide to look it up to figure out what it is. I believe this is a lab manual or some kind of manual, but in it the author states, “be a debating is accomplished by rapidly agitating a sample with a grinding medium (bead or balls) in a bead beater (device that shakes the homogenization vessel).” I hope this helps anyone else that was just as confused as I was when I read the words “bead-beating.” https://opsdiagnostics.com/notes/ranpri/OPSD_Bead_Beating_Primer_2014%20v1.pdf
I believe the purpose of washing the cultures before they are resuspended, is to make sure that they are sterilized before the next step in the procedure. I’m wondering what exactly the cultures are being washed with, unless I over looked it earlier in the paper.
What does finding the sequences and putting them in a phylogenetic tree do for the experiment? Does it just make it easier for the observer to see the difference sequences or is it used for something else?
Madeline I was thinking the same thing! I’m wondering what is exactly causing the Fla to be completely degraded by YE, but Nap cannot be degraded at all and Phe is slightly degrading with the presence of YE.
Why did the degradation of Fla decrease with the increase of its concentration? Is it because there wasn’t enough time to degrade a higher concentration or is there something else involved?
I find it interesting that this chart shows what each sequence is most closely related to, but also how closely they are related by percentage if I am understanding the chart correctly.
Is it the different concentrations of YE in different mediums that is changing the degradation of Fla? Or is it also because they are changing the concentration of of Fla?
In the previous paper we just read about how PAHs are pollutants in water, so I believe that the difference between coastal and terrestrial is that they come from different sources. But I am interested in finding out what else makes them different.
I agree that it would be very interesting if the same genes are responsible for degrading both marine and terrestrial PAHs, but if they have different sources would the same genes be able to degrade both?
What exactly does the M9 medium do? It contains ampicillin, thiamine, and glucose, but they do not explain what exactly they are using the medium for. Like lab this week certain tests are used to test for certain things, so what does the M9 medium test for?
I find it interesting that we have essentially done part of this in lab; where we put a culture on a plate and see if it has blue colonies. Its also interesting to see how the things we do in lab are used in different experiments and how we can relate what we have done to the experiments we read about.
Do both clones pH1a and pH1b both have the same 10.5-kb Sau3AI fragment? If so why only those two clones, if they got pH1a and pH1b from the genomic library wouldn’t all the clones they make from it have the same 10.5-kb Sau3AI fragment?
Why would the identity of the amino acid sequences be different if it is the same molecule? Are they different because they are using different alpha subunits or is it because of something else?
I find it interesting that they have less than 44% of their amino acid sequence in common. I also find it interesting that alpha and beta subunits had similar trees.
It’s interesting that PhnC is involved in the upper and lower pathways of degrading naphthalene. If it is involved in the upper and lower pathways of degrading naphthalene does that mean that it is more efficient in degrading it?
Christy, I was thinking the same thing. In an earlier paragraph it talks about how PhnA1 has alpha and beta subunits, so I’m wondering what effect of having one or the other or even both would have on their ability to degrade hydrocarbons.
I find it interesting that the oxyanions arsenate and arsenite are able to be a redox couple. And that the redox reaction that can help other organisms gain energy for growth. I find it interesting that Arsenic has different uses, besides it well known use of being toxic.
If As(III) and As(V) are found in periphyton, does that mean that the redox reaction that is occurring between them is helping the periphyton grow? That is what I’m getting from paragraph two if I am understanding this correctly.
I believe that the primers could have been the problem for not being able to obtain an authentic PCR. But could it have also been because of the enzymes that they used? After obtaining the PCR in lab and talking to Dr. Ni Chadhain she talked about how some enzymes could work for some genomes and not for others.
I find it interesting that As(III) and As(V) had opposite effects when being introduced to the dark regimen and the light regimen. Is this happening because it is showing how one can be an electron donor and one can be an electron acceptor and vice versa?
I agree that it is interesting that they came from the same biofilm, but have different temperature ranges. I’m wondering if this has an affect on their incubation experiments with being in the dark and light regimen.
After observing figure 4 its interesting that As(V) and As (III) have graphs that look very similar. As(V) and As(III) start out at different points but each graph looks similar to the other but inverted. I believe this is showing how one can be the electron donor and the other can be the electron acceptor in a redox reaction.
I find it interesting that even though acetate is important in anaerobic processes is was not found to have any stimulation on As(V). I’m wondering what could be that cause of this and why does this happen.
Jessica, I was wondering the same thing. I feel like they could have done the incubation experiments to make sure that what they thought was correct, but I’m not exactly sure.
While it is very interesting that organic and inorganic compounds can increase PAH degradation, how would this practically look in day-to-day life? Would we add these compounds to potentially contaminated foods? And how would we limit respiratory uptake? As someone mentioned above, I can’t help but wonder what the consequences would be in the environment.
Can it be assumed that Rhodococcus sp. is the better option when compared to the original strain of Rhodococcus? And between Rhodococcus sp. and inorganic nutrients such as nitrogen, which would be a more practical way of degrading PAHs in the environment?
After knowing the results of the study, I wonder what effect nitrogen or phosphorus would have on degradation of PAHs if used in conjunction with CMGCZ?
Since degradation was also examined in YMSM plates to determine the effects of yeast extract on the degradation potential of CMGCZ, I’m curious to know if MSM or YMSM plates supported higher rates of degradation. I think it will be interesting to see if the YE had any effect on CMGCZ.
Why was CMGCZ unable to degrade Nap in YMSM, but had a higher rate of degradation for Phe and completely degraded Fla? Is there a nutrient supplement that would support all 3 PAHs instead of just 2?
When looking at Fig. 2, the degradation of Fla in YMSM from days 1-3 appears to resemble the Fla degradation in MSM from days 5-7. It seems that YE in the YMSM plates shortens the time that it takes for Fla to be degraded. While I think the effects of YE on Fla degradation is interesting, I wonder what the effects of other inorganic and organic nutrients would be in comparison to YE.
Originally when I thought about the effect that YE had on CMGCZ’s ability to degrade Fla, I was only focusing on the fact that it completely degraded Fla and ignored the fact that it was unable to degrade Nap at all. I can now see where CMGCZ’s inability to degrade Nap would leave that PAH in the environment, whereas unenhanced CMGCZ at least has the ability to degrade a small amount of Nap.
Before Rhodococcus sp. CMGCZ is used for bioremediation purposes, I think more research needs to be done on it. More research could lead to a strain of Rhodococcus sp. CMGCZ that degrades Nap and Phe at higher levels instead of just Fla. Also, the effects that this would have on the environment, whether good or bad, are still unknown. Despite the need for more research, I think CMGCZ could prove to be a great option when it comes to degrading PAHs in the environment.
I think it would make sense for marine bacteria to do a better job at degrading PAHs in marine environments than terrestrial bacteria. I would like to know how Rhodococcus compares to the two terrestrial isolates mentioned here.
This study focuses on different PAHs than the previous study which makes me wonder if there is a difference in types of PAHs found in marine environments versus terrestrial environments?
Maybe I’m missing something, but what is the benefit to adding antibiotics? I thought this study was also focusing on PAH degrading capabilities of a bacterial strain.
Are there any disadvantages to forming the monohydroxylated forms instead of the cis-dihydrodiol forms? It says that the cis-dihydrodiol forms are the typical products, but doesn’t say if the monohydroxylated forms cause any problems.
Even though the PhnA1 amino acid sequence exhibits a high percentage of identity with the amino acid sequences of other bacterial strains, I wonder which strain is the most effective at degrading PAHs. I also think its interesting that PhnA1 exhibited 51-62% sequence identity while other alpha subunits exhibited levels of sequence identity lower than 45%.
Since phnA1 and phnA2 alone did not result in an active dioxygenase, but did show dioxygenase activity when coexpressed with phnA3 and phnA4, does this mean there is a link between dioxygenases and ferredoxin/ferredoxin reductase?
Just because PhnA was unable to convert anthracene and monocyclic aromatic hydrocarbons, could there be other genes in Cycloclasticus that could convert those hydrocarbons?
I wonder if these bacteria/archaea could provide any benefits to other arsenic rich environments besides hot springs, and could they use any other toxins as sources of energy?
I’m interested to know what role these arsenic metabolic activities might have had in the Archean earth, and how they have adjusted to all of the environmental changes since then.
Is it common for clones not to show any similarity with anything from the GenBank database? And why were the sequences that didn’t show similarity with anything excluded from further analyses?
I think its interesting that there were more types of Archaea that were found than Bacteria, but at the same time I would almost expect this due to the harsh environment they were found in.
I found it interesting that the temperature ranges differ based on if oxidation or reduction is occurring and whether or not they are exposed to light or dark. Why does the presence of light affect the temperature ranges for metabolic activities?
I think its really interesting that a new type of aerobic As(III)-oxidizing microbe could have been found in this system! I wish they had used an additional primer in this experiment so we would know if it was more likely that the lack of results came from using the wrong primer, or its a new microbe.
I think its really cool to think about how the processes that are being studied have been occurring for billions of years, even though the earth has changed so much since then.
Even though the Archaea are not involved in the arsenic cycling, I wonder what their role is in that environment? I understand that they are there due to high salt concentration, but what is it that they are doing?
I know that even the most remote areas of the earth are still affected by the actions of humans, but I did not know the extent of our presence in Antarctica. I also did not previously consider the effects on microbial communities. Usually when people think about environmentalism they automatically think about larger, well-known, and charismatic organisms such as penguins.
I appreciate the fact that the proposed solution to this problem is using the natural abilities of the bacteria that already exist within this environment. It’s almost like the earth is trying to fix itself and self-heal, and we just need to help the process become more efficient. I also wanted to mention I think that it’s really neat we are doing essentially the same experiement in lab as what is descibed here.
Both PCR and Sanger sequencing are extremely important experimental methods that changed science and allow us to do research like what we’re reading about in this paper. However, I did some reading to refresh myself on how these processes work, and I found that Sanger sequencing is actually considered more expensive and inefficient compared to next generation sequencing. This makes me question why they chose the Sanger method and if there are specific reasons as to why Sanger sequencing was used in this case.
I think the biosurfactant production is important to measure here because sufactants will help to break down the PAHs present in diesel oil, including phenanthrene.
The three highest phenanthrene-metabolizing strains were all taken from diesel contaminated soils. I wonder if this is because the bacteria there have been adapting to the presence of the diesel, and how long did it take for the bacteria to be able to metabolize diesel this efficiently?
It’s comforting to know that they identified a few different strains that are known to be useful for degrading PAHs, but that there are possibly more that just haven’t been found yet.
I did some quick googling to get an idea of what vector sequences, chimeras, and singleton sequences are. I vaguely remember learning about chimeras before, but vector and singleton sequences were new to me.
I remember learning about that experiment!
I understand that C13 isotopes are carbons with an extra neutron, but how are they accessed for use in an experiment like this? Are they collected somehow or can you create them?
Why are there fewer reports in (HMW) PAH- degrading microorganisms? Is it because they are harder to work with/to find? It is interesting to hear how these bacterium can degrade such a damaging neurotoxin. However, do these bacterium cause any negative effects to the environments?
It is really interesting to find out how this process works! I have done similar techniques in other classes but never really understood how the reactions worked.
If there is such a minimal percentage in the difference between the MSM and YMSM would have it been easier to exclude one of them in the beginning? Or would this be different depending on the bacteria inside the sample?
I’m curious if the different strains of degraders are used for different applications or if they all can be reproduced in the environmental remediation, pharmaceutical, and chemical industries.
I wonder if Rhodococcus is practical to use/obtain. I always hear of different types of bacteria but I don’t understand how easy it is to replication/obtain. Would it provide adverse effects to use it frequently?
If the product of Rhodococcus sp. CMGCZ is not reported in the ARHDs would this make the results only subject to this specific one? Have they tried replicating this experiment to see if they got similar results with different dioxygenases?
Upon reading this paragraph I was reminded of how in our last paper they experimented to see how these PAH degraders were able to digest PAHs within the Rieske center. I wonder if they will perform similar experiments here.
Will we have access to the tables in this study before we begin our papers? Also, I tried to look up Luria-Bertani agar plates but did not find results as to what their purpose may be in this experiment.
This paragraph made me think back to the first paper. I wonder what the amount of dioxygenase α-subunit genes were reported for that species of bacterium.
While interesting that they were not able to obtain the PCR products I was curious if these biofilms are the experiment or if they were made as a precursor to what they did.
How did they know that the fermentative alkalithermophile Anarobranca californiensis was isolated from that area of the hot springs? Was it tested before and they are assume nothing has hanged?
Why do we not see more organisms present in total? I understand that it is an extreme environment but I always assumed there would be thousands of different types in any given environment or is it typical for a few (6 in this case) to dominate an area?
Since these incubations were cultured in a sealed bottle would have been beneficial to the experiment to remove the waste products? or would it have contaminated the results?
I’m curious if other saltwater microorganisms in the ocean exhibit a lot of similar characteristics or if this lake is substantially higher in salt content compared to the ocean thus the same characteristics or environmental pressures wouldn’t be there.
Why would the temperature range vary? I expected both the As(III) oxidation and the anaerobic As(v) reduction to be productive at approximately the same temperatures. Is it simply the light-driven have adapted to addition temperature due to heat produced by the sun?
If it was determined that acetate amendment was not present in the As(V) reduction “implying that chemoheterotrophy did not drive reduction” then what would be responsible for the drive of As(v) reduction?
I’ve noticed in this paragraph and the one before that they are consistently monitoring and regulating the temperature of the samples. Is this simply standard practice or are PAH-degraders sensitive to temperature changes? If so is this sensitivity the reason there are not many variants of PAH degraders?
I am struggling to understand why they are performing these steps. I understand that they were isolating the PAH degraders but I was thinking that they had already done so in the earlier steps.
I am confused why they say that P.centronellolis does not have any reports of PAH degradation. However i’m the previous paragraph they mention that it showed good results in phenanthrene degradation.
I was unsure of what culture turbidity was. After some searching I found that it a way of measuring the amount of particles suspended in a liquid by using light
I was unsure if falcon tubes were a specialized type of use. Upon looking it up I found they were simply clear tubes that allowed you to see the sample.
I just want to make sure i’m understanding this section correctly… Since dna-Sip is a delicate process they preformed three variations of incorporating carbon into the biomass of the obtained sample degraders. This was for comparison of the results of carbon degradation and pah degradation that were run simultaneously?
So is this paragraph saying that in a experiment separate to the one in the previous paragraph Cycloclasticus was found in other parts of the sample core?
Since E.coli has the gene that is required for selenate reduction then the researches will not be able to use it as a control as they did in the last experience that, correct?
Why do the fused rings and absence of terminal surfaces in PAHs provide resistance to biodegradation? Is this simply because rings tend to be more stable compounds.
I would be interested in seeing a compare and contrast between the Nakheel beach region and Kuwait and Iranian coasts. I wonder how drastically these PAH degraders if at all in the Arabian Gulf region.
Does bioremediation present any ethical concern or issues? For example, the creation and use of genetically modified organisms because of the potential risks that may appear from altering a genetic code.
Are there any downsides to using bioremediation? Such as possible spread of these bacteria to other areas? How could you control where these bacteria go? Could they possible have a negative effect on an area which does not require bioremediation?
The CTAB extraction method is known to be one of the best methods for DNA extraction. Are there any downsides to using this method? For example, contaminants from plant tissues?
In reference to the N7 strain, what characteristic could be present to allow it to have maxiumum growth at 400ppm? That high of a concentration of NP created a drastic decrease in growth in all other strains but N7 exhibited maximum growth at these toxic levels. Why?
It is evident that the N7 strain has the highest growth in comparison to the other strains throughout the whole process. Regardless of the concentration, N7 is always showing more growth. But overall, the figure resembles the pattern of a bell curve, meaning there is a steady increase in growth – then a peak of all strain growth around 400ppm NP – then they all show a drastic decrease above that concentration.
What was the purpose of cloning the operon genes for the catechol meta-pathway and glutathione-S-transferase? Was this because they already knew those genes were useful in degradation and detoxification? Or that they wanted more present so they could further classify them?
Alignment of DNA sequences refers to the process of arranging these sequences to determine segments that may be similar to one another. This helps to further compare the strains of DNA and see which ones are similar as a result of structure, function, or evolution.
In the extension, or elongation, process of amplification is used to add a new DNA strand that is complementary to the template. This usually doubles the amount of DNA target sequences. Each time the extension/elongation step is complete there are more strands available to act as templates for the next round. Consequently, this leads to the exponential amplification of the target sequence.
Why did they choose to characterize this strain in particular out of multiple that were probably isolated? Did they have prior knowledge to this strain that determined it would be the best to further research?
Is there any significance to the strain being about to degrade via two different pathways? Salicylate and meta-cleavage pathway? Is is commonly found in PAH degrading strains?
Since there is no previous report on the meta-pathway gene of S. paucimobilis being able to degrade PAHs would this be the first discovery for this strain? Could this lead to the discovery of more bacteria capable of using this pathway to degrade PAHs?
What type of genetic modifications could they perform to improve PAH degradation? Would they be genetically modifying the strain to contain more meta-cleavage pathways?
Is the color appearance of the springs caused by the type of bacteria that inhabit it? Is the green appearance due to the cyanobacteria and red appearance due to the purple bacteria?
There is a big temperature gap between the storage of the the slurry sample and the biofilm for DNA analyses, why? The slurries were only needed to be kept at 5 degrees C and remained for several months. Where as the biofilm was storeed at -80 degrees C only until extraction.
That is correct. In the dark, anaerobic environment the Arsenate, which acts as the electron acceptor, needs an electron donor to be reduced to Arsenite. When no donors as present it cannot be reduced because no electrons to accept. Sulfide is a somewhat decent electron donor so it gets reduced a little bit. But H2 is the most favorable electron donor, to all of the Arsenate is fully reduced due to the abundance of H2.
In these environments, there are many microbes present so this slight decrease in As(V) (or increase in reduction) was due to abiotic processes of other microbes that may be around.
If i had to make an educated guess, the term “in vivo” means occuring within/inside another organism and “de facto” means that something is actual, or in fact.
So, for them to use those terms it would mean they are saying it does “in fact or in reality” occur “inside the organism”.
It is important to research which bacteria will do best in what environment to ensure the best results in naphthalene degradation. Is there a way to lessen the presence of other microorganisms that might compete with naphthalene degrading bacteria? Or should we look for other options for PAH bioremediation by observing the way nature breaks down cyclic aromatic compounds in similar environments?
Even though PAH bioremediation is considered effective and benign, if large amounts of carbon dioxide are released during this process then the possible ramifications need to be considered. High levels of carbon dioxide are seen as a problem in the environment as a whole and the adverse effects within marine ecosystems have already been noted. If PAH bioremediation was used how would we remove the excess carbon dioxide from the environment? Or would we continue to contribute to the threat of marine ecosystems and marine life? Other alternatives to PAH bioremediation should be considered and preventing more harm to ecosystems should be our top priority.
How accurate of an estimate does the turbidity measurement provide? Is there another technique that would provide more accurate data but still be feasible to preform? Does the calibration curve ensure that naphthalene is fully removed? How do you determine a calibration curve and why is it necessary?
What nutrients do the ONR7 medium provide in this experiment? How do you determine which medium would be best to use? What kind of benefit does the rotary shaker provide? Would bacteria still grow without the use of a rotary shaker?
What is the reasoning behind using 16S rDNA gene sequences that are available in public databases for comparison? Is this because of the availability or because these databases are found to be the most accurate? What is bootstrap replicates and neighbor-joining analysis? Why is this the best way to infer tree topology? Would there be a better way to infer tree topology even if it took longer or was less convenient?
What is the major difference between N1 and N7 strains when compared to N16 and N18 strains? What makes these strains more productive in degrading naphthalene? What is the GC-FID method and why is it more feasible to use to analyze microbial growth when compared to other methods?
What kind of marine environments are best for isolation of naphthalene degrading bacterial strains? What makes some marine environments better than others?
Even though some of the genera can uptake crude oil, are there any consequences of it doing so? Does genera produce any harmful effects in the environment?
Is microbial transformation and degradation also thought to be the best method for removing other pollutants from the ecosystem? Which strains are the ones that can degrade PAHs completely and what are their limitations?
Why was there a need to clone the catechol meta-pathway operon genes and glutathione-S-transferase gene? And what was the reasoning behind thinking this strain would be good for performing bioremediation of PAH pollution?
What is the point of disrupting with 99cycling of sonication for 3 seconds? What differences are identified with a molar absorption coefficient for CDNB and GSH?
Are the tests that were done typical for bacteria characterization? Are these the main tests that most use to identify bacteria? Are there any more tests that should be considered and why?
Is the meta-pathway present in all bacteria able to degrade PAHs? Or does the meta-pathway make it more efficient? How would understanding the meta-pathway facilitate the improvement of PAHs degradation through genetic modification?
How are clone libraries constructed? Were groups containing one or two more clones selected because of time consideration? If not, then what was the reasoning behind it?
What is the reason that all aoxB clones failed to show similarity to anything in GenBank? Why is RFLP analysis the way that the clones are grouped? What makes this analysis the best?
Why would this cyclic phenomenon only show in oxic/anoxic experimental biomes? Isn’t it necessary even in completely anaerobic ecosystems? How does the cyclic cycle differ from that in anaerobic ecosystems?
Is it more likely that the primers are not suited for the environment or that there is a novel organism present? What ways would we determine what kind of mechanisms the novel organisms use for As(III) oxidation?
Through oil spill is very critical, I personally still believe that nuclear leak should be the most critical environmental pollution. in 2011, there is a case of a nuclear leak in Fukushima Daiichi, Japan. I heard that it will affect the coast and living organisms for over 50 years.
But this paragraph is to introduce bioremediation, I may not really put questions on the giving background.
I think the yeast plays the role of catalyst, it is used to speed up the reaction and they may want to set up the experiment to test PAH removal in different concentrations and bacterias. it is easy to lower concentration by adding solvent. The no substrate 100 ppm PAH is more likely a control group or starting material.
In figure 2, 2 samples are incubated at 21 degrees, but I think in the deep sea, the bacteria should habitat in a much colder environment like what material and methods page mentions 4 degrees but the method page also mentions 21 degrees. Is that mean those bacteria in the deep sea can grow at room temperature and function similarly in both temperatures? if so, can we find those hydrocarbon-degrading bacterial somewhere else like the crude oil on the surface of the sea?
Why proving that fnr gene can solely active selenate reduction result in only fnr gene? Is it possible for ogt gene can also solely activate the selenate reduction? In figure 2, it also just shows the graph about proving the function of fnr gene, is there a technique issue for testing with only ogt gene?
I think it states that E.coli specifically contains selenate reductase because of the non-oxygen environment in the intestinal which does not need the oxygen-sensing transcription factors to detect the absence of oxygen and regulate selenate reductase activity like other cells with oxygen. in other words for aerobic cells, the selenate reduction needs a transcription factor to regulate. But I don’t understand how the hypothesize given the bacterium adapts from aerobic to anaerobic.
If it is a catabolic pathway it seems it would be more prominent around higher concentrations of PAHs. Could an increase of the concentration of microbial degraders offer information about the environment? Thinking more in marine environments. Or even soil? It may be easier to test for the actual PAHs.
The genetic acquisitions of Alteromonas sp. SN2 is that referring to horizontal gene transfer and is it possible to use genomic analysis to identify which genes have been transferred?
Maybe PAHs aren’t the sole source of carbon, or these microbes have the ability to degrade and not necessarily be completely dependent on them? Then the mutants could still be viable. I’ve started working with some arabidopsis mutants that have a disrupted continuum of cell wall, plasma membrane, and actin cytoskeleton leaving them struggling to produce much cell mass. They are pitiful compared to the wild type, but enough for our purposes.
P73T isn’t the only gene responsible for degradation then. Alis is right it would not survive in this environment or have metabolites for that matter. It’s interesting to see where this is going. How complex is the pathway and the mechanisms involved?
The possible potential on P37T for use in oil spills is exciting. It is quite remarkable to learn about how many genes have been linked to HGT, and to consider how many more marine microbes possibly hold similar potentials.
This is making sense to me(hopefully). It would be like looking into a (arabidopsis) mutation of actin depolymerizing factors (ADF) and testing strains of ADF3, ADF4, and ADF5. Systematically testing slight variances to get at precise mechanisms of functions.
I’m very excited for our 16S rRNA gene comparisons in class. Personally I can find clarity in concepts more readily when performing tasks, or at least gaining a visual for the process in its entirety.
I wonder if the novelty of the P73_0346 has anything to do with it being a marine bacteria. In the introduction it was stated that proposed metabolic pathways for degraders have been rarely seen in marine bacteria.
My understanding is that COG annotation is representative of speciation events, which in bacteria happen quite rapidly. So the database was constructed to show genetic differences between closely related species, I think it may highlight HGT?
This is why PAHs accumulate? They are hydrophobic therefore not readily susceptible to non-metabolic degradation. Making them “persistent organic compounds”(intro)
How do scientists determine which naphthalene-degrading bacteria to introduce into the environment? If these bacteria are not able to compete with other microorganisms in a certain environment, how do scientists determine what to do next for naphthalene degradation in that environment?
Were the time frames between each assessment of the growth curves of the isolates the same each time? Also, could a different assessment routine have shown better results?
I recall the BP oil spill where so much oil was dumped into the ocean that the animals were coated and the coast wasn’t safe to swim in anymore. This is what sounds like the result of us beginning to travel and explore the Antarctic more will be. We see the major effects these events have on macro-organisms but pay very little attention to what happens to the microorganisms.
The more natural and protected we can keep Antarctica the better because there is a lot of potential for studying bacteria that has been frozen in the layers of ice there. Not only to mention the native species only found in Antarctica, the fact that only native microorganisms can be introduced is a relieving statement as dumping so many foreign organisms into the water would surely have an effect on the native species.
This is very cool to see that the experiments with the microorganisms being done here could possibly lead to a way to degrade oil in the future. This, if done correctly could lead to ways for us to help restore areas that have been damaged by oil spills.
Typically PCR is used for DNA replication but I believe that is only for a segment or certain strand. They were likely trying to dilute and isolate a certain colony of bacteria that could use a specified fuel source. Once they isolated the bacteria however, I do believe that PCR would have been a plausible option.
Were the bacteria collected with the syringe the ones that were to be isolated for further replication? That is what I had gathered from this as it seems that the bioattractant was used to separate the desired bacteria from the other bacterium.
I recall that as well, it seemed that the values were from 25-250 listed in the manual. I am unsure of this but I think that the video linked in the pdf said you could count up to 350 and that would be an acceptable number.
In this experiment the first time the bacteria replicated it took much longer to replicate, such as with a 48 hour lag phase, but in the next culture it didn’t take near as long. I am wondering why it took so long for the bacteria to replicate, the first time especially. The culture mediums would have had a large amount of nutrients that the bacteria could digest, and normally in rich media bacteria replicate rapidly.
That’s what it seems like from the previous posts. I believe it was in the introduction that the idea of dumping bacteria into the ocean to remove oil spills by bioremediation was suggested. I would imagine that the same idea or process would be used for this, though in this paragraph in the second sentence it states that Antarctica is a region that introducing forbidden material is forbidden so that may hinder the process or the research here could at least be used for alternate situations.
I am wondering id they will be able to use this research in Antarctica. In the second paragraph it was stated that foreign materials can’t be introduced into Antarctica, so does that mean this research is only for finding bacteria to be used in other situations?
That’s what it sounds like to me. I would imagine that the oil bunched together and got too dense to float on the water so it eventually descended to the ocean floor.
Since the bacteria are psychrophiles would this be the highest temperature they could tolerate, and since bacteria grow better at their upper range of temperature tolerance then wouldn’t that mean this would theoretically be the best temperature for maximum bacterial growth?
They probably froze them so that the bacteria wouldn’t replicate and essentially become a culture broth. These bacteria are psychrophiles so they would likely excel in replicating if they didn’t completely freeze them.
I think that is likely the answer. The PHE is probably the most similar to PAH and there was likely a mutation allowing in the enzymes/proteins that allow for the degradation of PAH.
I understand that the acid inhibited bacteria are used as a control group, but why are they using acid controlled bacterium specifically. Wouldn’t they simply be dead bacteria?
I imagine so, unless this was the maximum temperature the bacteria could tolerate since bacteria tend to metabolize and grow more at their maximum tolerable temperatures.
The 13C model was the only one being observed in the study since the 12C and the 14C were just mainly for comparisons it seems. So if they weren’t being studied then I imagine they were only concerned about results from 13C.
It seems like the As(V) is the electron acceptor and As(III) is the electron donor since they are listed as a redox couple. It also seems like they have isolated chemoautotrophs as well as photoautotrophs meaning yes, they will use CO2 as carbon sources.
I would imagine that this was were they decided to do research on this bacteria in Japan or that they did in fact find that the soil in Japan had a higher concentration of Arsenic making it more hospitable to these bacteria.
I am slightly confused as to how they adjusted the pH to 9.3 using HCL as it is an acid. I would think adding an acid to a solution would make it more acidic rather than making it more basic.
That sounds to be likely what happened, either that or the research on these primers were conducted in conjunction with the research on the bacteria in the biofilms.
Does tis mean that these bacteria are functional or micro anaerobes since adding oxygen nor removing it effects the rate at which they degrade materials?
Would this mean that this is possibly a new form of lithotrophy that hasn’t been identified yet, and is likely specific to Arsenic at these oxidation states?
What makes the yeast enhance degradation? Could there be another co-substrate that could be added to enhance degradation where the PAH ppm doesn’t have to be lowered?
It is weird they found a unique PAH-biodegration pattern and never isolated it or investigated it. What could have caused this and why was it only in one location?
Was there a difference in the PAH when the oil spill originally happened from after they did PAH degradation? Did the PAH degradation make a major impact?
With the concentration of extractable PAHs in the sampling locations high, what is the normal range? Do the phosphorus and nitrogen levels have any correlation to why the PAH is high?
Is there a reason to why all the isolates had different colony morphologies but were still rod shaped and gram-negative. Would there be a difference if they were gram-positive versus being gram-negative.
Why was there such a big difference in temperature between the core collected in November and December? Does the temperature difference have to do with the well developed Beggiatoa mat?
Hi Melanie! Great question I was wondering the same about what clearing zones where. I understand now that when adding the PAH it makes it cloudy and when it is removed it turned clear.
Hi Tessa I totally agree with you that its very interesting to see how this could go in so many other directions. It’s great to see how this work added to the knowledge we know about hydro-carbon degrading bacteria.
Hi Anna I was also confused on what effect new were suppose to be seeing since the PAH was removed. Thank you Dr. Chadhain for clearing up my confusion.
Hi Madelyn I was also confused as to why E. coli S17-1 was not able to reduce Se(VI), but E. coli S17-1 combined with cosmid clone E. coli pECL1e was able to reduce Se(VI)? Does this have something to do with being grown in the minimal salt medium?
I think it’s extremely interesting to see how different bacterial species can degrade various toxic chemical groups in similar manners. I am definitely curious to see the next few sections on this paper to see how their experiment compares to that of the PAH degraders.
I was interested in reading exactly where PAHs come from and how they are released into the environment, and what I found was that these chemicals are found naturally in coals, tobacco, wood, and other materials that release PAHs when burned (sited below). The “harmful effects” listed in this paragraph also seems indicative of smoking cigarettes and second-hand smoke, which are both ways that PAHs may enter someone’s body.
(https://www.epa.gov/sites/production/files/2014-03/documents/pahs_factsheet_cdc_2013.pdf)
The efficiency of the catabolic processes of the rhodococci to transform these harmful PAHs into TCA cycle intermediates is extremely fascinating. It seems that these bacterium play a crucial role in degrading environmental contaminants for human health and also in the nitrogen and carbon cycles for maintenance of atmospheric conditions.
I’m noticing here that these researchers used MSM plate media, just as we did in Lab 2 to begin isolating our naphthalene degraders. The composition of MSM seems to be commonly used to enrich the growth of the PAH’s.
After performing the initial steps to isolate our naphthalene degrading bacteria in lab today, I am curious to start to see results and draw connections between our lab and what these researchers are finding. As someone stated before me, perhaps incubating for 2 weeks will have more of an effect than incubating for 48 hours – I believe we will be able to observe and record the differences seen among the time frames in our own lab work in the next few weeks.
The results of the degraded samples on the MSM (for Nap and Phe) are contrary to what I thought they would be. I drew a parallel between the researchers using MSM in this experiment and our class using MSM for our soil sample in lab to a significant contribution in the degradation of PAHs. It is also interesting to see how significant the degradation of Fla was on both MSM and YMSM plates compared to the other two samples.
This makes me curious about what our MSM and R2A plates in our own lab experiments would look like if we observed them over a period of 8-14 days, instead of 2-5 days. I checked my group’s plates 48 hours after the experiment and only had one plate with blue/black colonies. It will be interesting to view our plates on Tuesday and see if we can make any connection between our results and this experiment.
When I was reading the “Results” section on the efficiency of CMGCZ to degrade the different PAHs, I was very interested in reading that Fla (HMW) was degraded 100% after 8 days on the YMSM but Phe (also HMW) was not significantly degraded after 8 days. The report mentioned in the “Introduction” that there have not been many reports on degradation of HMW compounds, so I found it peculiar that a HMW compound used in this experiment was degraded entirely.
I wondered the same thing. One would think that a smaller, aromatic structure would be more easily degraded than a larger aromatic. In addition to there being such a striking difference between the Fla and Nap degradation, also throw in the comparison of the Phe degradation. While Phe is also a HMW compound, the degradation more closely resembled that of Nap (LMW) than Fla.
My interpretation of this experiment based on the “Materials and Methods” section was that the researchers were changing the concentrations of Fla in the medium and recording how efficiently the CMGCZ would degrade the various concentrations. This experiment was performed on both MSM and YMSM mediums.
As we read further into this paper, it will be neat to draw comparisons between the Cycloclasticus here and the Rhodocuccus bacteria from the last paper. It looks now as though degradation of petroleum is a major role of study of the bacteria is paper 2.
It is interesting that this paper places more of an emphasis on the effects that PAHs have on marine ecosystems, rather than terrestrial (i.e. human) ecosystems. Perhaps the researchers wanted to study effects on marine life that could possibly have an effect on terrestrial lives through seafood, water, and other possible ingested pollutants.
An expression vector is a plasmid or virus designed for gene expression in specific cells. In this experiment, I presume pPhnA and pPhnC are plasmids that will be inserted into dioxygenase genes.
I would think that a defined medium would be used to eliminate the number of variables affecting the growth of the bacterial species. If it were a complex media, it may be more difficult to isolate the colonies needed for the experiment.
I found by researching online that LB agar plates are a common complex media used in microbiology research. I presume that the purpose for using this kind of agar is to allow growth of multiple forms of like (E. Coli and the recombinant plasmids).
In lab 6 that we completed this week, we also used a type of endonuclease to digest the nuclei of our naphthalene degrading isolates. It is starting to become easier to draw connections between our lab work and the work performed in these research experiments!
It is not directly stated, but I am assuming the aromatic oxygenase gene is the gene responsible for degrading PAHs? The introduction of the paper does not specify a particular gene they are searching for, just that Cycloclasticus sp. should have degrading capabilities.
I am extremely puzzled by this figure. Each of these structures (A, B, C) are physical maps of genes on the plasmids – I understand that. However, I don’t understand how restriction sites play a role in the ORFs? I can not deduce why they are shown in the figure or their significance.
I am a little stumped on how this paragraph says “the order of [genes] was found to be quite different from that of analogous genes reported previously” as well as “A5 harbored no plasmid… localized on the chromosome.” Are the researchers implying that they found something new with their research, or that previous accounts were incorrect regarding the location of these genes? Or are they simply pointing out that they found different results?
I’m noticing the term “cluster” being brought up quite a few times throughout the Discussion. What exactly does this mean? For example, “it falls outside the major cluster”; I’m not sure how to interpret this.
That’s a very good point! I never considered the idea of a PAH degrading bacteria to be categorized as an extremophile, but for some reason an arsenic-degrading bacteria does seem like it might fit that category.
I’m just wondering here, why did they want to place the samples on an artificial hot spring medium? I would think they here is when they would want to determine specific composition and abilities of their bacteria, so why wouldn’t they use a selective media?
So, I’m just making an educated guess here, the radioassay is what the researchers are using to determine the strength of the arsenic-degrading genes in the bacterium? Because they already know that these specific genes are present based on the genomic library construction, this portion of the experiment is now testing those genes?
This table is interesting because it shows more percent identity with species of archaea than with bacteria. I guess because archaea are generally less-widely studied, it surprises me that the majority of their matches were with these species.
It is so interesting to see that there are opposing effects that As(III) and As(V) had in the dark versus in the light. I would think that this is happening as a sort of oxidation-reduction coupling, where both reactions must take place to keep the cycle going.
Reading this discussion is definitely helping me connect a lot of the dots we saw in the figures in the Results section. Where this paragraph states “which implied the involvement of physiologically different anaerobes” seems to make more sense to me in regards to the differences in the dark and light degradation of arsenic seen in the last section.
It is interesting here that they did not find the results about acetate and chemoheterotrophy that they were expecting to find. Are they stating that the strains of interest capable of degrading arsenic are not chemoheterotrophs? Or are they simply saying that acetate is not an electron source for their metabolic functions?
I think this is really interesting too. When studying for this last exam, I remember reading something about how it’s easy to study the 16S rRNA gene because it is “highly conserved and universally present” among various types of organisms. Pretty neat!
Lameace, I too am curious to know how they would go about re-testing this since the first set of primers was unsuccessful. Would another type of primer work? Or would it be better to use a transcriptomic approach, such as a gene chip microarray? Is there a benefit to using one over the other?
Since PAH exposure occurs via respiratory uptake and some of their effects seem to deal with the respiratory system, does that mean they generally occur from instances where smoke enters the air, like smoking cigarettes or burning stuff?
So after reading the paper in its entirety, I’m getting the vibe that Rhodococcus is a key player in degrading PAHs, and that it is one of the few bacteria that can degrade HMW PAHs. Are HMW PAHs worse than LMW PAHs?
Hey Alexandria! I was wondering the same thing you were and did a little research to find that Rhodococcus can degrade a lot of things! Some interesting things I found that it could degrade was estrogens and alcohols.
By using the different types of media, are the researchers testing to see which offers a better source of growth for the PAHs? Also, why were two separate stock solutions of 1% Fla made? What purpose does spraying the agar with Fla made in acetone have?
I was wondering the same thing Lameace! I know that PCR is used to create copies of the DNA sequence of the target molecule but I’ve never given it any thought as to whether or not another gel could be used. Like you said, maybe a certain gel is used for a certain purpose and the researchers used the 0.8% Seaplague GTG agarose for a certain function.
I am thinking the same thing you are Lameace. I would think the washing of the bacteria would help eliminate any potential contaminants to the sample and allow for the culture to be pure.
Compared to R. erythropolis in MSM, the R. erythropolis in YMSM degraded at a faster rate in the first few days but seemed to catch up in a significant drop off by day seven. It even states in paragraph 7 that there was a delay of degradation in the MSM as opposed to the YMSM, which was delayed for the first 24 hours. I’m wondering what media, then, would be most beneficial to use given their seemingly different rates of degradation but overall similar effects?
So this may be a dumb question but I had no idea that biologists or molecular microbiology used aspects of organic chemistry, like proton NMR or IR to analyze compounds! I could also be reading the information and chart wrong, and it is some other kind of analysis, but either way, it looks really cool!
Based on the cluster information from the Rehmann et al. (2001) study, it seems Rhodococcus sp. CMGCZ falls into the second set. Although it was able to use other PAHs besides Fla, it definitely favored that PAH in particular.
With the finding of the Rieske [Fe2-S2] center, do you think it could potentially serve as a better degrader of Nap, Phe, Fla, and ILCO or limit Rhodococcus sp. CMGCZ’s already established success? Personally I would love to see further analysis into it.
This introduction is very similar to the first paper we summarized. It seems like the main difference is in the testing of a separate bacterium from Rhodococcus.
So this study’s primary focus is more so on marine PAH degraders than terrestrial ones? I wonder what aspects of marine PAHs make them unique from PAHs found on land.
I never thought of it like that until now! You’re totally right though, Tara, I wonder how much harm we are actually doing to our ecosystems in the name of maintaining it and allowing controlled growth? The grilled meat releasing PAHs is sad too, looks like I’ll be baking my chicken from now on.
I’ve always wondered what motivates a researcher to choose the type of analysis and tools of measurement they use because I can imagine that there are many different ways. How do they know that the one they chose will be the best fit for their intended study.
Is it a requirement for all researchers to report their nucleotide sequence data into these genetic libraries or is it a sort of professional courtesy?
Even after watching the video describing the cloning process and going over it in class today, this process is still very confusing to me. I am starting to understand bits and pieces of the process however which is good progress in my eyes.
Is there any significance to the fact that PhnC activities were similar among all catechol compounds? Also, does 3-Methylcatechol having the highest activity give it some sort of preference for further testing?
Does a dioxygenase not being able to use a certain substrate, limit its potential or ability? How is PhnA dioxygenase effected by not being able to use anthracene?
I found it unique too because it may suggest an entirely new group of genes that are similar yet distinct to the ones we already know about. I can imagine that alpha and beta subunits can often share similar phylogenetic trees having close to the same function in protein folding.
Tara I was wondering the same thing about the implications that this ability could mean for us. I know that arsenic is deadly to humans so us being able to use a bacteria’s/archaeon’s natural ability to utilize it could help our survival against it.
I was wondering the same thing Nolan. I thought it was noteworthy that the researchers even used the word “surprisingly” as if they also expected to obtain a PCR in the first place.
This may sound dumb but what is the purpose of the researchers going into full detail about the appearance and location of the hot springs they used in testing? Does it allow for future researchers to find as similar samples as possible or does it allow there to be no loose ends or grey areas in our current researcher’s methods?
Sarah Grace, I would also love to see a phylogenetic tree based on the arsenate respiratory reductase genes to potentially see the other bacteria and archaeon that’s able to use arsenic as an energy source.
Based on figure 1, it looks as if As(V) and As(III) are inverses of each other in the light to dark reduction of As(V) and the light to dark oxidation of As(III). This trend kind of makes sense to me in that for most redox reactions, there’s an equal reduction for every oxidation. The paragraph below suggests that both the reduction and oxidation of As(V) and As(III), respectively, are capable of cooccurrence.
The finding involving Ectothiorhodospira is good, right? Kulp being able to find an isolate of the genus that’s able to grow by photosynthetic oxidation of arsenite in an anoxic environment suggests that the 16S rRNA gene of the studied bacteria is similar to that of a known arsenite user. It was also interesting that the archaea genes were more diverse but they all seemed to originate from hypersaline environments (harsh conditions common of archaea).
Hey Sarah Grace, I thought it was cool too of how the rate of cycling was diminished. Your comparison of it to our own aging processes is very unique and comforting that we’re not the only organism to always be slowly dying lol.
What is the significance of the different suggestions of the arrA clones? Like the ones that compared closely to Ectothiorhodospira strain PHS-1 vs. the ones that didn’t indicate what biofilm microbes were associated with them?
Hey Tim, I was wondering also how effective transcriptomics or bioinformatics would be in a situation like this. I wasn’t sure where either could be applicable, but learning about these testing methods in lecture has made me wonder where they can be suited.
Hey Hang, that’s a super interesting concept to think of because I would think that activity could be present and similar results replicated in a different environment, following similar conditions. I would also think that controlling for those conditions in a new environment, especially one that is open and very dynamic as opposed to a lab setting, would be incredibly difficult.
No studies have investigated PAH-degredation from the region mention in the Arabian Gulf. Why is this? With studies in other areas, such as, the other sides of the Arabian gulf and the Kuwait and Iranian coasts, wouldn’t it be beneficial to add the data from this region to those studies and view/study it comprehensively?
Would light and scanning electron microscopes be the only valuable microscopy for this experiment.Would transmission microscopy be a advantageous tool for the experiment’s results or would it contribute little to the interpretation of data?
The article says that low concentrations of N and P are present and the environment has a high toxicity, which might limit productivity. How would it be definitively proven that is the N and P concentrations have a limiting effect and not the toxicity alone. Also, going off of that, how would one alter the concentrations and toxicity in order to reverse the inhibiting effect?
The last sentence says that OD is not suitable for larger size PAHs as a growth assessment method. If this is the case, why was implored? Is it because initially size of the PAHs were not known?
It is mentioned that O. intermedium, P. citronellolis, and Cupriavidus taiwanensis are less researched in regards to PAH degradation. I can’t help but wonder if there is a reason for this or if it has just been overlooked until now. Moving forward from this experiment, do you think that more researchers will take interest in these strains?
I agree with Danielle’s comment, PAH degradation seems to be under-researched. I have to wonder if this is due to financial limitations such as not enough funding or if it is a lack of interest in the topic, or something else. It seems as though this topic could be a major ecological boost for many ecosystems.
It seems to me the goal of this research group was to only identify and describe the different types of PAH degraders in the Guaymas Basin. Will research of this nature eventually lead to the cultivation of PAH degraders in areas where oil-pollution is high?
At the end of the paragraph, it is mentioned that the the relish spot are oil droplets. Is this color difference due to the high concentration of the oil in that specific spot, also is sites with that high concentration of oil targeted?
Madelyn, I had the same thought. If this is true, shouldn’t the two sample have different traits in order to make it possible to be present at these deep depths? And if variations how present, does this mean the two samples are actually both cycloclasticus?
What is the disadvantage of carbon13 being distributed among other members of the microbial community? does it skew results or make it difficult to interpret results?
Hi Melanie, I had a similar question. What causes the difference in color between red or white? Were the colonies grown on the plates similar to the ones we use in lab (blue colonies/white colonies) or is this color a characteristic of the actual bacteria?
The first sentence in this paragraph states that it was hypothesized that the FNR transcription factor regulates selenate reductase activity. What might other possible regulation factors be?
The last sentence of this paragraph states that fermentative growth on glucose allows for reduction to occur while anaerobic growth on nitrate inhibits it. Would a possible expansion of research be to determine whether the nitrate itself is limiting the growth or whether the anaerobic growth is?
This first paragraph does a great job at identifying the problem and explains why this topic is important. When discussing oil spills I usually think about the effects on more large and notable animals (such as ducks or small fish). However, these environmental pollutants harm every aspect of aquatic life down to the micro biome
I think it’s really cool that scientist have possibly found a natural way to reduce pollutants in the soil. I understand that no foreign bacteria could be introduced into the soil seeing this could easily make problems much worse. I am curious on the possible outcomes of increasing bacteria that’s not very plentiful in the soil. What are the possible negative effects of this on the micro biome?
I’m not going to act lol a fully understand everything happening here. But i do find the process of PAH quantification to quite interesting. It so different from any quantification method I’ve ever used in a lab.
Here the authors do a great job at pointing out the possible sources of error. They noted their bias to the three strands of focus in their research and noted how that could have negatively effected the outcome of the experiment.
Although a lot more needs to done the results from the experiment seem to be very promising. I’m excited to see how this will be applied in a real world setting.
Was there a specific reason those 3 contaminated sites were chosen? Would it benefit to collect a sample form farther into the gulf rather than 2 from an island and 1 from a shoreline?
From 200-400 ppm, N1 had a greater growth than N10. I thought it was strange that at 500 ppm, the N1 and N10 strain were at equal growth, and at 600 ppm, the N10 strain had greater growth. No other strain experienced a change like this. Is there a specific factor that caused this change?
As stated in paragraph 9, N7 had BATH of 73.57% which was almost 7% higher than any other strain. Is this cell cur face hydrophobicity a key factor in why this strain had the highest level of growth even though N1 had the highest percentage of naphthalene degradation and E24%? Would the N7 strain be favored for bioremediation?
What exactly about the structure of a gram-negative bacteria makes it able to tolerate PAH uptake better than gram-positive which has the strengthening agent peptidoglycan? Is it because the fatty acids are bonded through the amine groups from glucosamine phosphate, or is it because of the different functions of the proteins in the peripplasm?
In the results section, I made the assumption that high cell surface hydrophobicity must be a stronger factor than emulsification activity because the N7 strain was more successful in growth than the N1 strain. I see now that there is a direct relationship and both are important in choosing the best naphthalene bacteria strains.
N7 belongs to Sphingomonas. Does the fact that this strain was identified and reported to use naphthalene as a soul carbon and energy source have anything to do with why the strain was so successful in growth?
I had the same question as Menesha, and also wondered if the binding abilities of teichoic acid embedded in the cell walls of gram-positive bacteria was a factor for its acceptability.
I had the same question as Menesha, and also wondered if the binding abilities of teichoic acid embedded in the cell walls of gram-positive bacteria was a factor for its acceptability.
Are specific PAHs amounts concentrated in similar areas? Such as a certain PAH amount is in areas with high forrest fires, and a level of concentration is in areas that are industrial dense?
Are specific PAH amounts concentrated in similar areas? For example, is there a certain/ similar concentration amount found in areas with high forrest fires, or a certain concentration amount found in industrial dense communities that are similar?
I had a similar question, and looked up the recipe to this media. Apparently it is easy to prepare and provides a broad base of nutrients. It was formulated for studying lysogeny in E.coli and has even been referred to as “Lysogeny Broth”.
Ampicillin is not a part of the LB recipe. I know that it is an antibiotic that can be used to treat bacterial infections, but why is this specific penicillin antibiotic used?
I thought it was interesting that the activity was similar in the PhnC’s that were fused and unfused. Does fusing not have an affect on the activity usually?
I had a similar question. Would an experiment with the same variables yield the same results with PAH degradation on chromosomes rather than the plasma?
I do think that it would conserve resources to have one pair to partner with many different deoxygenates. However, keeping conservation out of mind, would the experiment benefit from making specific ferredoxins and reductase for each deoxygenate the cell would encode?
I remember reading that reductase and ferredoxin were 2 components that were normally needed for electron transfer to the terminal dioxygenase, however, could there have been other enzymes used for this electron carrier system?
I had the same question and looked up that the diel cycle is a 24hr period where the chemical and biological structure of a microbial can change as a consequence of changing light intensity. Changes can be detected using microsensors in pH, H2S and O2. Detecting the rate of these changes reveals zones of greatest microbial activity.
RFLP is a technique that was one of the first techniques used for DNA analysis, however after looking into it, I noticed that it is not used widely anymore. Why was is used for this particular experiment and was there an alternative that could have been used that would have been more modern and accurate?
Because the results were not clear, are there separate/ alternative tests to determine if the oxidation activity was due to the aoxB-independent mechanism, to rule out that it was not due to the inefficiency of the primers?
I also agree with their approach. I also agree with the fact that they started off with As(III) in higher concentration to be more efficient. I assume that they experimenters knew it As(III) would oxidize in light and would therefore get results faster.
Responding to my previous comment, this makes me wonder if the results would have partially differed if they would have started off with As(V) at a higher concentration in the light phase.
Was there a certain amount of time that each population was exposed to the specific temperature? Does the amount of time that it is exposed have anything to do with the result of the temperature range experiment?
Using a non-oxygen acceptor allows chemolithotrophs to have greater diversity, while sacrificing energy production. Sometimes the use of an enzyme is needed for oxidation. Would the use of one change the results of this reaction?
[“Oil contamination can generate detrimental changes in soil properties, including modifications in maximum surface temperature, pH, and carbon and nitrogen levels (Aislabie et al., 2004).”””Altogether, this results in a significant decrease in species richness and evenness, and a large decline in soil biodiversity of contaminated soils (Saul et al., 2005; van Dorst et al., 2014, 2016).”] Mentioning species richness, the number of species within a community, and species evenness, the commonness of species within a community, it is also equally important to discuss species diversity, that of which combines species richness and species evenness. Biodiversity is more than just the counting of species, but it rather encompasses the genetic variability among organisms within a species, the variety of different species, and the variety of ecosystems on Earth.
[“This process can be achieved by promoting the growth of endogenous metabolizing bacteria in contaminated sites (biostimulation) or by directly seeding contaminated sites with pollutant-degrading bacteria (bioaugmentation). Since the Antarctic Treaty impedes the introduction of foreign organisms into the Antarctic continent, bioaugmentation can only be implemented by the use of native microbes.”] Why is it that only ‘native microbes’ are allowed into the Antarctic continent? What effects would the use of ‘foreign microbes’ have on Antarctic’s contaminated soils?
[Briefly, non-polar compounds were extracted from culture media using two volumes of hexane and vigorous mixing for 60s.]
What is the reasoning behind using ‘two volumes’ of hexane? Did this make the extractions more concentrated thus increasing the excitation-emission spectra of each sample?
[Cultures were grown with agitation at room temperature for 5 days, and phenanthrene metabolizing strains screened by change in medium color from clear to yellow due to the generation 2′-hydroxy muconic semialdehyde, a degradation product of phenanthrene (Treccani, 1965).
After reading the line above from the text, I became interested in the degradation product of phenanthrene, 2′-hydroxymuconate semialdehyde; that of which, I soon discovered is ‘formed from catechol by the enzyme catechol 2,3-dioxygenase during the degradation of benzoates’ & is ‘hydrolysed into formate and 2-oxopent-4-enoate by 2-hydroxymuconate-semialdehyde hydrolase.’
Since Chemotaxis seems to not play a part in PAH degradation, does that mean the bacteria treated the PAH like every other degradation process? If that’s the case, is there a particular reason it failed to differentiate PAH?
Since 16srRNA identified the isolates of Rhodococcus erythropolis, S. xenophagum, and Pseudomonas guineae, why then was only Pseudomonas guineae unable to process the diesel fuel as an energy source? Why is it that all three strains are psychrophylic yet 2/3 were able to utilize diesel, would they not all be able to utilize diesel in that case?
Dayana, I agree! “Increase in temperature increases the rate of oil degradation by bacteria” makes sense considering bacteria are more likely to thrive in warmer conditions. Warmer environments such as farms, industrial sites, landfills, and/or onsite sanitation systems are bacteria-rich and would likely be beneficial when using a microbial bioremediation approach.
Abraham, similar to you: I was also curious about ‘biosparging.’ After a quick google search I found: “Biosparging refers to air injection at pressures and flow rates sufficient to deliver supplemental oxygen, but less than those required to volatilize significant contamination. Evidence indicates properly designed sparging systems significantly enhance both biodegradation and volatilization.”
I was unsure as to what ‘siderophore aerobactin’ was, but after googling I now know that ‘siderophore aerobactin’ is a bacterial iron chelating agent found in E. coli & is commonly found iron-poor environments, such as the urinary tract.
“Following incubation overnight at 30^C, V. fischeri ES114 colonies harboring transposon insertions were selected by growth at room temperature on LM agar plates containing erythromycin.”
Knowing that erythromycin is an antibiotic, is there a reason they chose this specific antibiotic as opposed to other antibiotics?
Ryne,
I asked the same question in a previous paragraph as ‘erythromycin’ was specifically chosen & I am curious as to what influenced their decision regarding the antibiotics used within the experiment.
“A strain of Vibrio nereis encodes 6 receptors, but it cannot produce any siderophores (Thode et al., 2018).” Why is it that a strain of Vibrio nereis cannot produce siderophores?
“Moreover, both vibrio species produce a variety of public goods including extracellular proteases, chitinases, and QS autoinducers, all of which can be monitored in real time.”
Because I was unsure exactly what a “chitinase” was, I google searched to discover that “chitinases” are enzymes that degrade chitin & contribute to the generation of carbon and nitrogen in the ecosystem. Chitin is a component of the cell walls of fungi and exoskeletal elements of some animals (including mollusks and arthropods).
“It connects the recycling of organic nitrogen to the ultimate nitrogen loss from the oceans, because its products are substrates for denitrification and anaerobic ammonium oxidation (anammox), the only two presently known nitrogen loss processes.”
Anammox (anaerobic ammonium oxidation) is a reaction that oxidizes ammonium to denitrogen gas using nitrite as the electron acceptor under anoxic conditions. Anammox was an important discovery in the nitrogen cycle.
“It is responsible for the formation of the large deep-sea nitrate reservoir.”
Are hydrothermal vents the only cause of the surplus of nitrate in deep ocean water?
“A 20- to 40-m-thick suboxic transitional zone, characterized by low oxygen and undetectable sulfide…”
Due to the low oxygen and undetectable sulfide levels in the Black Sea, does life still exist within it? Are these levels toxic?
“Abundance of total Archaea was taken as the sum of cren- and euryarchaea.”
In efforts of focusing on the data collected rather than ‘how’ they collected it, would a high point within this paragraph be the equation: “[cren-] + [euryarchaea] = total Archaea abundance”?
In this paragraph, I feel as if this particular sentence holds importance: “15NO2 − production was measured in the same anoxic 15N incubation vials as in the anammox rate measurements, but 15NO2 − was analyzed as N2 after a two-step reduction by acidified sodium iodide and then by copper at 650°C.”
From the Figure 1 graph(s), I was able to gather that as ammonium levels increase, nitrate levels decrease. Additionally, the high anammox bacterial levels are indicative of nitrogen loss during oxidation.
From the Figure 3 graph(s), I was able to discover a correlation between the species of bacterial and crenarchaeal amoA expressed & the depth of the ocean; the species expressed falls as the depth rises.
It is really interesting that Diesel Oil is most commonly used fuel in Antartica when Antartica is considered to be one of the most untouched places on Earth. So to read that we are using one of the most toxic fuels there is disheartening. But I am glad that they are trying to help combat using other toxic things.
I am interested to see what their outcomes were especially because the antartic ecosystem is particular different than a lot of other common ecosystems in the world. Also the latest source from this paper was 2017. I would be interested to see what it is now in 2021. Have we made process with this? Or are we still harming our environment?
WOW! It is incredible that one could access the DNA sequence code online, and the fact that you could find something similar is amazing too. I also find it interesting that it took so much to get this approved/ submitted. I wonder if the process is this intense now? Especially because what they used was from 2000.
I wonder why after two months they were analyzed? Is that so they were for sure isolated and not contaminated? Does that happen a lot in other experiments? I understand it is coming from Antartica, but that is still an extremely long time.
It is really interesting that they are psychrotolerant rather than psychrophylic, when you think they would do better because of their hostilely cold environment. But it makes sense that it was seen previously because bacteria can get trapped in ice.
It is really interesting that it has to be under the right conditions to grow. One could see that they had a huge struggle with getting D43FB to grow in paragraph 8. But I am intrigued at how picky these things are by deciding what special environment they want to grow in.
I totally agree with you! I never would have guessed how many specific bacterias there are in soil. But also just in this experiment alone where they had what seems like an endless opportunities to find almost any bacteria. It is really hard to wrap my brain around how many bacteria there truly is.
I wonder if this is true with extreme environments in the ocean/space, or if this is just by land? Why do they have to be extreme environments, and what is the benefit? Also it makes one wonder that yes, what they are researching is amazing but is there an easier/ cost efficient way to travel to these environment and take samples?
This is incredible that bacteria is abundant. This is insane that these are responsible for recycling of the hydrocarbons. Especially because these things are not even on the surface.
How many other bacterias are there and how much does this cost? Also, how does this help us in the future because finding more oil-rich sediment is great but how long until this taken to use for resources? I think I understood this correctly, if not, can someone help me understand better?
does pressure play a part in this? Does it matter how they keep it in lab? No pressure has to be applied to the tubes? Even though this bacteria is found at extreme depths?
I am interested to see which temperature works because 4 degree C and 21 degree C are two completely different temperatures. So I’m curious as to why so high and so low temperatures to test. Is there no in between?
I wonder if the amount of oxygen had something to do with why they could not cultivate. Bacteria at the bottom of the floor verse in the shadows uptake and use different oxygens. So, maybe the cultivating has something to do with how much oxygen they get.
Referring to the last paragraph as well, I am curious to see if the shallower bacteria degrade oil like the deep sea because I know oil floats but I also see oil in shore too as well from spills and what not. So I would like to research more on that too.
Interesting that the process of nitrification and denitrification affect it so much. Especially because we do not hear denitrification getting used that often.
I have actually drove past this lake when I was in California, I had no idea about the springs and what not.This is so crazy, it was crazy in person too!
I am wondering why they failed to show significant similarities when it seems like they would. I wonder what would happen if the testing did go on further?
They talk about using the heavier DNA, but then acknowledge the fact that the lighter DNA could have the reaction to degrade it too? A lot of these ending results experiments seem to pick the better one but then note that the other data not used could be it. Who is studying this other weaker experiment? Will it get studied?
Nothing was mentioned that said that these naphthalene-degrading bacteria were introduced to the Persian Gulf. Does that mean that these species are native and the purpose of this study is to identify those that thrive in these PAH- and oil-rich environments to further study them, or is the purpose to identify which species that they introduced was performing best and in which environments?
Is the PAH bioremediation really environmentally benign? Granted, it fixes the pollution problem, but doesn’t it create a dead zone (which is an oxygen-deficient area) from the formation of CO2 and H2O (from both the breakdown of the PAHs and presumably the use of O2). Also, the creation of the microbial mass would be similar to algae blooms in that when those microbes die off, it creates a large food source for the decomposer, which use O2 to feed on that microbial mass, thus also creating a dead zone.
What is the purpose of doing the estimation of the remaining naphthalene when a more accurate GC measurement was taken later in this methods section (paragraph 11)? What does this version of the measurement tell you that the other does not?
It was mentioned earlier that the significance of emulsification activity and BATH test determine how well a bacteria can attach to a carbon source. I can understand the surface hydrophobicity (because the bacteria’s hydrobicity can be used to attach to the PAHs hydrophobic region), how does emulsification affect the bacteria’s ability to attach to the PAH source?
So when these scientists discover these strands, even though they are showing the general genus and species that they are related to, is this the equivalent of someone discovering a new subspecies of a known animal? Also, how different can different strands of the same species be?
If they are trying to determine a bacteria’s ability to break down PAHs, why are they using naphthalene as the determining food source? Shouldn’t they use more complex molecules with more benzene rings to make it more selective since PAH stands for polycyclic aromatic compounds, which indicates that any multitude of benzene rings could be present in a single compound. If they just base their research off of naphthalene doesn’t that mean that the more complex segments of the PAHs could be left over?
The Gram-negative is what I would have hypothesized to do best because it has that outer-membrane. Why, then, did the last experiment conclude Gram-positive were the naphthalene-degraders? Were there no G- present in that environment, or did they just not break down naphthalene? Also, did the G+ degraders have special adaptations that allowed it to tolerate these types of environments without the outer membrane? What could some of those adaptations be?
I thought there was not much difference in the ability to break down a simpler PAH such as naphthalene compared to the more complex PAHs that they are talking about, with the only difference being that it takes longer. If that’s the case, then why are they distinguishing between naphthalene-degraders and other, larger and more complex PAH-degraders?
How are these genomic sequences able to determine whether this strain is a good candidate for performing bioremediation? Are these sequences compared to known sequences of organisms that are capable of degrading, or do they say something about the speed and/or ability of this strain to perform the necessary catabolic pathways to break down the complex PAHs?
Is there a way to separate out the enzymes to determine what each one does specifically in order to identify the ones involved in the PAH break-down? Or do we have to just look at the activity results from a solution containing all of the enzymes and just be able to say that at least a few enzymes are present in the cell that are proven to break-down PAHs?
Why are they looking at the plasmid DNA sequence? If the genes encoding for PAH-degrading enzymes are found in the plasmid, does that mean that this bacteria doesn’t naturally degrade PAHs, but rather got these sequences from another organism present in the soil (and maybe even from on of the other organisms from the original soil sample)?
They are testing for all of these different aromatic compounds, but how many different types of PAHs can actually be found in oils, plastics, or other potential PAH-contaminants? And, just out of curiosity, on average how many aromatic rings do they have?
So is the purpose of determining if two ORFs (such as phnH and phnI) are detected next to each other to see if they could encode for proteins that are part of the same biosynthetic pathway? Or is it for another reason?
What does it mean by CDNB-accepting? Caroline and Marissa said that GST was use for detoxification, so does that mean CDNB is a toxic component of phenanthrene, or is it a toxic by-product of the breakdown of phenanthrene, or does it indicate something else?
Is there a reason that they needed to compare both sequences? Was it because the 16SrDNA sequencing isn’t accurate enough, or did they just want to make sure they classified the strain correctly?
Why are they focusing on meta-pathways for degrading PAHs? Is there an advantage to having that sort of pathway compared to another type like ortho-cleavage pathways?
Just a thought, but, does the ability of bacteria and other unicellular organisms to undergo horizontal gene transfer affect the accuracy of determining phylogeny based on genome sequences, since they can transfer some of their genome to others not necessarily of the same species or genus?
Is it more common to find As(V) reduction and As(III) oxidation coupled between different species or coupled within one organism (as in one organism can do both)? Also, is this paper going to focus on the relationship between an As(III) oxidizer and an As(V) reducer or on a single organism that can do both?
I know hot springs (which is created through volcanic activity) also contain H2S in addition to the As(III), and I know there are many microbes that use H2S in the process of chemosynthesis. Since these microbes use the As(III), does that mean that there is a selective advantage to using As(III) for photosynthesis rather than H2S in chemosynthesis?
Why would the assimilation of acetate not be stimulated by the addition of arsenic oxyanions in light conditions if it is the reduction of As(V) that helps drive the oxidation of acetate? Shouldn’t more acetate be assimilated if more As(V) is added?
Can you really say that the PHS-1 strain only does the oxidation of As(III) to As(V) if the PHS-strain is only measured for the As(III) rate rather than the the As(V) reduction rate (since the graph only indicates that under the light reaction that this oxidation takes place and does not consider the As(V) reduction)? Shouldn’t this strain be cultivated in the dark to truly determine that the reduction if As(V) does not take place in this strain?
How do they know that the rates of the oxidation of As(III) and reduction of As(V) decreases over time? Is there another study that they are referring to?
How did they not know which microbe was able to perform the As(V) reduction if they were able to isolate each Bacteria or Archaea strain (or did they really isolate each strain?) and test them on their performance in a dark environment? Couldn’t they just measure the amount of As(V) that was reduced in the dark environment to determine the rate of reduction?
I find it interesting that we are exposed to PAHs through ingestion; however, substances such as glucose, yeast extract and broth aid in the degradation of PAHs. What foods contain PAHs? Is it public knowledge that these foods are possibly harmful?
I agree Madeline. I am very excited to see our results as we isolate and characterize naphthalene degrading bacteria from local soil samples in the lab!
I find it interesting that this study used a soil sample from an abandoned oil field in Japan, while we used a soil sample from the base of a light pole in Dr. Ni Chadhain’s neighborhood. I guess this just shows the abundance of PAHs in the environment.
I was curious about this initially, as well. However, after reading the entire article, I realized that the added yeast extract in YMSM plates increases the rate of PAH degradation.
I was curious about the clear zone formation technique. I looked it up and found that the clear zones around the colonies represent its ability to inhibit other microbial growth.
If obligate marine bacteria are possibly more significant PAH degraders in costal marine environment rather than terrestrial bacteria, why is there only a limited amount of research? It seems that this could be substantial to the health of marine life and, therefore, human that consume seafood.
I never really considered purchasing the chemical and reagents. I guess I just assumed that the researchers obtained and/or isolated them independently. So I found this interesting!
I find it fascinating that we are able to clone specific genes in DNA and use them to our advantage! I wonder if this could be a permanent solution for the PAH crisis?
Louie, I’m a little confused by this as well. I reviewed my notes from class yesterday and noticed that I wrote “primer walk” following the direction of the arrows down the nucleotide sequence.
I wonder why PhnA was not able to convert anthracene and monocyclic aromatic hydrocarbons, but is otherwise able to degrade a wide range of aromatic hydrocarbons.
Spencer, I too was curious about the function(s) of ISP. So I did some research and found that they fulfill functions in electron transport, enzyme catalysis, homeostatic regulation and sulfur activation.
It is interesting that this paper, in contrast to the first two, studies the characteristics of both bacteria and archaea. I am curious to see how the remainder of the paper will be presented.
Hi Derria. I was a little confused by this at first too, but I looked it up and it seems to mean a 24-hour period. Here is the definition I found: “of or relating to a 24-hour period, especially a regular daily cycle, as of the physiology or behavior of an organism.”
Jesse, I was curious about that as well. I think that the sequences that did not show similarity were excluded because they would not be useful in the remainder of the experiment.
I was also wondering if the times of the sequencing were significant. The gap between the first two months was only two months, but the gap between the last two months was seven months.
I find that interesting as well, especially since we’ve primarily studied bacteria thus far. I also suspect that the harsh environment contributes to the larger number of archaea vs bacteria.
I find it interesting that the genera of the nearest neighbors of representative clone are so diverse, because the top 10 matches on both of the BLAST searches I performed in lab were all from the same genus.
Unfortunately, with any highly visited area there is always going to be pollution affecting the natural habitat of the area. It seems like these explorers do not have any consideration for the environment that they want to visit. Maybe a solution would be to restrict the amount of people that come to Antarctica per year and put rules in place to designate what kind of equipment that can be brought and where transport of vehicles is allowed to minimize the areas of oil spillage.
It is very amazing to see how scientists can use the bacteria from the Antarctic soil to degrade a key component of diesel-fuel. This allows to them to help fix some of the damage that has been done from the oil spillage. I wonder if these scientists have looked into using electric vehicles as an alternative to transport and if the effects are less detrimental. As long as these diesel-fuel trucks are coming through at high volume, it will be hard to tackle to effects of oil spillage.
This makes me wonder what environmental bacteria was collected that is unique to Antarctica. Due to the extreme weather conditions, I imagine that the microbes collected would be very different compared to the ones collected in the climate experienced in the United States. This also poses the question that if the environmental bacteria in the freezing temperatures of Antarctica are unique, are they more susceptible to the negative affects of diesel fuel?
The crystal violet solution and the use of an alcohol sounds like they could be doing a gram stain on the bacterial isolates. However, in this case, crystal violet was used to measure absorbance and assign the correct density of each culture. Is this another way of saying they used a gram stain to determine the thickness of the peptidoglycan layer, or is this another analyses altogether that just happens to also use crystal violet and an alcohol?
It’s interesting to read how scientists are able to observe if chemotaxis occurred. Instead of having to view motility under a microscope, it seems that they were able to determine the ability of chemotaxis by seeing if the bacteria were present closer to a target compound, where they previously were not. This paragraph gave me more insight on how lab processes are done.
The finding that the presence of heavy metals reduces the degradation of diesel fuels raises more questions about the environmental pollution. Is it possible that the traffic going through Antarctica leaves traces of metal from the vehicles and equipment along with leaving traces of diesel fuel? And if so, will the use of these bacteria be impactful on the diesel fuel or, instead, become a failed attempt?
This paper taught me that scientists are able to modify bacteria to enhance their capabilities. It seems extremely advantageous that this is something possible to further degrade the diesel fuel and ,in turn, help the environment. Could formation of biofilms, secretion of biosurfactants, or the ability to form chemotaxis be enhanced to enable the bacteria to work more effectively with the presence of heavy metals?
This paragraph is excellent at explaining the way that these scientists cultured the bacteria and how it can be further researched. Hopefully this paper inspires others to experiment with different culture techniques and will enable them to possibly find more types of PAH degrading bacteria. The more research that can be done on the different bacteria found in Antarctica, the more of an increased likelihood that the diesel fuel pollution problem can be overcome.
It’s interesting that scientists are able to spray on the growth substrate which then leaves a layer on the agar for the bacteria to use. This seems like a quick and efficient way to ensure the bacteria have their source of carbon that is evenly distributed.
It’s interesting that scientists are able to spray on the growth substrate which then leaves a layer on the agar for the bacteria to use. This seems like a quick and efficient way to ensure the bacteria have their source of carbon that is evenly distributed.
I wonder if 11 days in lab time is considered long or short to wait for sufficient results. Also, is there some type of enzyme that can be added to 13C to make the degradation and mineralization process faster and more efficient?
Now that the scientists have found that Cycloclasticus are obligate PAH degraders, it would be interesting to see how a scientist would use this information to enhance the environmental conditions of the sea floor. Are there bacteria similar to Cycloclasticus in other parts of the sea that are affected by oil or would these areas benefit from having scientists plant the founded PAH degraders in parts of the sea floor other than Guaymas Basin?
Looking at the figure, I’m assuming that all of the PAH degrading bacterial strains isolated are from the same ancestor. I wonder if the related strains from GenBank that are given would have the same or better PAH degrading properties? This might be good supplemental research to pursue.
Is the rusty color produced from some type of by-product from the chemical reaction taking place? Also, is this by-product harmful to the environment but just less harmful than oil that is not degraded?
When recreating the hot spring water for incubation, it says that O2 free N2 was dispensed in anoxic tubes. So, it’s safe to assume that the hot springs created an oxygen-free environment for these bacteria and archaea to live in? Or is this just for the purpose of the experiment?
I looked up crimp sealing and couldnt find an exact definition. I’m guessing it is some specialized seal on the serum bottles to keep the gases and samples inside and any microbes or dirt etc outside to keep the samples just as they were when collected. It also might be important to use to keep oxygen from interfering with these anaerobic microbes.
Shelby, I was wondering the same thing. I would also assume light could withstand a higher temperature. Maybe its possible that they needed a more specific temperature with light to be able to use it as an energy source.
It’s interesting that both As(III) and As(V) have almost the same optimal temperature and general shape of curve. The difference is that the dark incubated biofilm has a larger range of temperature to be able to oxidize as well as a slightly higher oxidation rate for each point than the light-incubated counterpart. Due to the close similarities, I wonder if the oxidation is actually the same for each mode of incubation, but possibly there are other aspects of the experiment that allowed for the dark-incubation to have a slightly higher oxidation rate.
I wonder if at 50 degrees C and over these microbial populations start to die off. Maybe the temperature that the biofilm is active is the window for which the biofilm can survive.
I wonder at what point did evidence of oxygenic photosynthesis arrive? What microbes that utilize oxygenic are the closest in relation to these arsenite degrading archea and what mechanisms do they endure that are similar?
I like how the introduction of this paper talks about how oil-degrading bacteria has been explored much more recently and that the sea floor would benefit from this type of research considering the oil spill in the Gulf of Mexico. I think we rarely think about the long term effects from the oil spill and that there’s still clean up to be done. I’m glad that research on oil degradation is being done in this regard.
It’s interesting how scientists are able to studying these hydrocarbon-degrading microbes with the use of SIP. I wonder where else these specific bacteria live other than off the coast of California and if it is possible to place these bacteria in different areas of the sea that they are not originally found to degrade oil.
The prior papers studied phenanthrene degraders as it relates to oil pollution and could be environmentally useful to understand the bacteria. For this paper, I understand that they are studying how the bacteria and archaea utilize concentrations of the different forms of arsenic, but what was their driving factor? Was it just for the sake of understanding it better or to use this research as a way to degrade arsenic when needed?
In the photosynthesis dealing with water and sunlight that we normally learn about, water is the electron donor and light is the form of energy. Anoxygenic photosynthesis is photosynthesis without water. So, these specific bacteria are using As(III) as their electron donor in place of water?
From this paragraph, it seems that the researchers received the opposite result that they were expecting regarding the samples’ ability to degrade hydrocarbons. They explained this, however, by attributing sample 4571-2’s lack of hydrocarbon-degrading ability to its use of sulfide instead of oxygen, an adaptation to anaerobic environments.
By using the picture and graph, we can interpret that there is one gene that is more abundant than the rest. That gene was in fraction 6-10, which means it represents 13^C heavy DNA, indicating that 13^C heavy DNA is found in high quantities.
After reading this paragraph, I was reminded of a personal assumption I made that in nature, bacteria lived closely to bacteria that have similar genetic makeups (offspring stay close to parents). It is interesting to me that bacteria that differ so much genetically, observed in their ability to degrade PAH and possibly morphologies, live in such close proximity.
I am interested in the reasoning behind the -20 degrees Celsius temperature i which the agar slants containing isolated strains were kept in. Was this to prevent colony growth and reproduction, as bacteria grow and reproduce more quickly at a warmer temperatute?
What did the phylogenetic tree tell the researchers about the axenic isolates? Did they’re evolutionary relationship to certain bacteria suggest a reason as to why it is the result of pyrene being used as a carbon source?
I am interested in whether the Gram test was positive or negative. I would also if the physio-chemical properties of the isolate’s cell walls play a role in determining what PAH the bacterium use as a carbon source.
Does BC1’s relation to O. intermedium suggest that it, too, effectively degrades high molecular weight PAH such as pyrene and benzo(a)pyrene? BC1’s relation to Brucella melintensis also suggests that it may be present in livestock and humans; could this information be applied to health or agriculture studies?
Isolates LB and LC are clearly different in that LC is a hydrocarbon degrader and did not grow in the PAH while LB degrades citronellol and grew wonderfully in the PAH. What about being a hydrocarbon degrader made the LC so incompatible with the PAH environment, or what about being a citronellol degrader made the LB so compatible with the PAH?
The basis of petrochemicals are hydrocarbons, which explains why this environment for ideal for studying hydrocarbon-degrading bacteria. However, once those petrochemicals are broken down, what are the recycled into?
It appears to me that the SIP technique has been utilized for identifying microorganisms and assigning them to phylogenies based on their ability to absorb a stable isotope. This does seem like it would be very useful if one of the goals of this experiment is to identify the number and diversity of PAH-degrading species there are in the samples.
As shown in the captions of the pictures, one was taken at the sampling site for background cores while the other was at the site for hydrothermal cores. What is the difference between these two?
I recognize SIP as a technique used in the last article to distinguish bacteria based on their ability to degrade hydrocarbons. It seems that certain sequences of DNA in these samples were identified by SIP, suggesting that these samples help the bacteria in biodegradation.
It is very intriguing how the enzymes in the different bacteria, which are made based off genetic code, could affect the SE(0) product structure. I think I understand this correctly, but the reduction mentioned in the first sentence is referring to the use of selenium oxyanions as terminal electron acceptors in anaerobic respiration right?
As there are two mutagenesis methods we have discussed, I wonder which these researchers picked from. I believe the best one to use would be site-directed because on of the objectives of the experiment is to identify important genes.
The LB medium that the bacteria was originally grown in contains many nutrients that promote bacterial growth, while the minimal salt medium would only allow the growth of bacteria that could use the included compounds as an energy resource. Transferring the bacteria to this selective medium was intended to isolate the target bacteria, correct?
If I am reading this correctly, the wild type of the cloned DNA was Km and Gm resistant, so when plated on LB agar supplemented with Km and LB agar with Km&Gm, growth on the first but lack of growth of the second indicted a mutation. Would that be a mutation in the Km-resistant gene?
I believe that is correct, that the minimal salt medium would separate the types of bacterial colonies by only allowing the certain bacteria to grow. Those that grow then produce red as a result of the selenium reduction.
A knockout mutation is one in which a gene is made inactive. This shows that any protein or function exhibited with that gene and then consequently not shown without the gene could be controlled by that gene; however, these researchers have to address that another reason might explain the lack of exhibition.
Since the regulatory genes from other Sphingomonas strains that degrade different PAHs have been isolated, would it be possible that these regulatory genes could be similar enough to give some insight to finding those associated with acenaphthene degradation?
From my understanding, it seems the gene cluster that was isolated is located on the same segment as the arhA genes, but these are not all of the genes used in the process of degradation of acenaphthene. The rest of the genes are most likely scattered.
From my understanding of previous papers and discussions, the initial oxygenase enzyme is the one responsible for the production of indigo from indole. So, if they have lost the ability to produce indigo, it could be due to the absence of the gene/gene product.
From my understanding, I think they are highlighting the structural similarity of fluoranthene to the other PAHs as being the reason that their biodegradation pathways are similar, not necessarily the same.
Celeribacter indicus P73T is the first fluoranthene degrading bacterium found in the family Rhodobacteraceae. They are studying this bacterium because of this and I think they are using fluoranthene because it is a good model PAH when studying PAH metabolism.
Does analyzing the intermediate metabolites of degradation of fluoranthene give the researchers information towards which kinds of pathways are being used, depending on the types of intermediates?
From my understanding, the increased percentage of genomic DNA encoding transporter genes in strain P73 is a result of the strains ability to degrade PAHs.
So, they were able to determine the pathway through a combination of computational predictions and metabolite analysis? Other than the metabolite analysis and the gene deletion, their entire approach has been computational predictions, correct?
Would the research approach they used here be considered a preliminary attempt to answer the kinds of questions the last few papers have answered? For instance, this group analyzed the genome and proposed a possible metabolic pathway, but did not perform any further testing to prove the proposed pathway.
Before reading this paper, I wasn’t very informed on this topic. I have learned a lot about the toxicity of PAHs. The statement I found most interesting was that the catabolism of aromatic compounds in rhodococci leads to the transformation of these compounds (found in PAHs) to TCA intermediates. I wonder if we could somehow manipulate this ability and think of PAHs in a positive way?
Based off the comment, “Fla…has potentially carcinogenic effects,” I am curious to know how much of a role it has in cancers studied today? The word “potentially” also stands out to me because I wonder if it hasn’t been studied enough to draw legitimate conclusions or if this statement is only relevant under certain conditions like temperature, part of the body etc?
Hi Jesse! Yes I think that it is safe to assume that Rhodococcus sp. is the better option with regards to this experiment because it neglects to mention otherwise. It also states, “the capability of Rhodococcus sp. to degrade high concentration of Fla does not exhibit homology with catalytic domain of previously reported ARHDs.” I interpreted that statement to mean that it was a better option!
I’m not entirely sure, but I think that it is a way to “purify” a sample of bacteria. Maybe it could be a way to control for unwanted growth during the experiment?
I’m not entirely sure, but I think this could be a way to “purify” the bacteria. Maybe the bacteria is washed to control for unwanted growth during the experiment?
I wonder how running the PCR product on agarose gel helps us to confirm apt purification? As someone who hasn’t run a PCR before, I wonder if that a “normal” way to conduct an experiment similar to this one or if this gel was chosen for a different reason by the experimenters.
I think this part of the results section is one of the most interesting. It’s amazing that we can use DNA to try and identify the different bacteria strains in a media. I think those percentages are pretty high and demonstrate good accuracy in the identifications.
The extent of differences between these three PAHs was interesting to me. I assumed Fla would be degraded more than Nap and Phe (which happened) but the individual percentages and the increase in residual PAHs for Nap YMSM surprised me.
I agree Dawson! I am also interested to know how popular rhodococci is as an organism in biology studies today. In other words, is this is a common organism molecular microbiologists use in research? Is the sentence, “rhodococcus is one of the most promising groups of organisms” a “given” in molecular microbiology research?
I agree Justin! I also find the sentence “there are studiess reported where bacterial strains degraded HMW PAH but not LMW PAH to be very interesting. I would have assumed the bacterial strains would be able to degrade lower molecular weights to an easier extent. I wonder what caused this to happen? Was there a sort of affinity to the HMW PAH that was lacking in the LMW PAH?
I think that this part of the experiment was really cool especially because we have discussed the lac promoter in class. Also, further research in our ability to manipulate genes in order to become over-expressed (or repressed) could have really interesting effects on various genetic diseases.
From what I understand, the ability to oxidize indole to indigo means that there’s a presence of an aromatic oxygenase gene. Kind of like in lab when we saw cells turn pink when placed on the Macconkey agar. The color pink meant that they could ferment lactose. So, I think that that means our degrades have that aromatic oxygenase gene as well. I interpreted that statement to be causal.
I agree! I think that this paper describes really cool techniques that give us the opportunity to learn more about metabolism and microorganisms in general. I also wonder what the not continuously clustered genes mean in terms of degradation potential of PAH, especially with the mentioned high degrees of similarity with the sequences used for catabolism. In transcription with regulatory genes order matters, so I assume it’s the same concept for this.
I agree! I also find it interesting that both genes function similarly despite the different order of the phn genes. Based off what we have discussed in class so far, I would have assumed that the order of the genes would affect the overall function.
I find it very interesting that this paper resulted in the possibility that PhnC is involved in both the upper and lower pathways for degradation of nap, phe, and biphenyl. This seems like a very different result and approach to researching PAH degraders than our first paper.
I think that the topic of this paper is really cool because not only are there bacteria that can survive in the presence of high concentrations of arsenic (a known toxin), they also the ability to use it to help increase energy. I think it shows the truly diverse and “smart” nature of microbiology.
I think the focus of the examination of the cycling of arsenic under anoxic conditions is very interesting and important. Due to the Earth first being anoxic, this will truly help us get one step closer to understanding how organisms survived through the use of cycling of arsenic in that time period.
I agree; that sentence stood out to me the most. I would be interested to see how the researchers responded to the lack of the PCR product and how it impacted what they chose to do next.
That surprised me as well. In classes we never mention bacteria possessing that same characteristic of archaea. I’m interested to see if there are any differences in their respective ability to live in environments rich in arsenic.
I am curious as to why singlet clones are not generally used for sequence analysis. Is there a lack of their ability to be sequenced because their RFLP groups are represented by only a single clone?
I am interested to see what the radioassays of the chemoheterotrophic or chemoautotrophic processes resulted in. I am curious to see the difference between the two groups of organisms with relation to arsenic biotransformations.
I am curious to know if the experimenters knew that the thioarsenic intermediates were soluble at high pHs before the they saw the errors associated with the sulfide-amended samples. I am assuming they didn’t but maybe they did and the use of high pHs was the only way to get the samples?
While the observation discussed in this paragraph is important because it shows which organisms are the dominant arsenic cycling ones, is the detection of only one clone type a substantial find? Are there usually more clone types when conducting experiments such as this one?
I found this part of the paper to be very interesting. It’s really cool that accumulation of As(V) allowed a new set of niches to open. I also find it interesting that the cyclical reactions are able to show a shift between aerobic and anaerobic conditions even though it occurred such a long time ago.
I would assume that the next step would be to try a different set of primers because that seems like an “easier” solution than trying to isolate new types of aerobic As (III)-oxidizing microbes and then doing follow-up biochemical investigations. However, I think if the oxidation was being carried out by a novel mechanism, that the new information obtained could be interesting and provide scientists with important information that would help us understand the way systems work more fully.
This paragraph specifically mentions petroleum PAHs. Were they chosen to be studied for a reason or does the bacteria have stronger and more diverse degradation effects on petroleum PAHs compared to others?
I agree Lane! I also wonder how this investigation could effect the marine-located PAHs that humans have contributed to. This experiment could open up lots of research and discussion about how to potentially minimize the damage that we have done through our use and disposal of petroleum products.
I agree Anna! I think that their environments have helped them become more equipped to degrade marine PAHs than terrestrial PAHs. I wonder if the difference in marine and terrestrial physical and chemical environments (such as internal temperature) could have something to do with optimal PAH degradation activity as well.
Tara,
I had similar thoughts on somehow isolating the specific ARHDs and then researching their functions as it relates to the catalysis of PAHs specifically. Perhaps having the ability to garner this material would prove to be a more efficient manner of degradation.
I suppose that also brings about some other concerns such as financial implications, as you mentioned, and also the potentiality for negative impacts on surrounding ecosystems.
I, too, found it to be interesting that the addition of particular organic and inorganic supplements could potentially amplify the impacts of PAH-degrading microorganisms.
I’m curious to know whether or not some of those supplements are more or less effective than others, and if so, what is the reasoning behind that? Also, perhaps there is a particular supplement ratio (a certain amount of nitrogen and phosphorus) that would create a highly conducive degradative environment. Is it possible that in the presence of certain amounts of organic or inorganic materials there would be a negative result, as well?
I’m curious to know the reasons why Rhodococcus is more versatile in its ability to metabolize a broader range of compounds.
Is there a particular structural difference between this strain and the others mentioned prior, such as Mycobacterium, that render it more effective on this front?
I am curious of the significance of the development of chromarods in hexane and hexane/toluene. In what way do these two materials together influence this development? Does this mixture somehow help further isolate the desired material similar to the chemical used as an anti-fungal in lab 2?
What significance does nitrogen have in this particular experiment as a “carrier gas”? I’m assuming it is used for the GC, but is it more efficient than other gasses, or is it the only gas that won’t denature the extracted material?
I think that the degrading properties present in MSM is particularly interesting; slow degradation followed by a bout of rapid degradation seems to be something of importance.
Perhaps this type of degradation could be more beneficial in certain circumstances and for certain bacteria?
I’m also curious to know why the level of degradation is diminished with an increasing Fla concentration.
Although there is still successfully degradation, the relationship between residual Fla and the concentration of Fla is one that I would like to learn more about.
What about concentrations below 100 mg^-1? Would there be a faster degradation or maybe an unwanted/unexpected result?
Since the relatively low amount of degradation of Nap and Phe was contributed to “enrichment and continuous subculturing”, I am curious if there would be a way to remediate this by adapting the technique.
For example, what would the percentages of degradation of each respective PAH be if the enrichment and level of subculturing was altered?
Would these changes potentially enhance the PAH utilization capabilities of Rhodococcus sp. CMGCZ?
I think its so interesting how different strains here are more or less effective at degrading particular alkanes/aromatics, etc.
The last point mentioned about the degradation of diesel by these two bacterial strains makes me curious to know if there would be an impact by adding both strains together.
For example, would the treatment of both R. erythropolis T7-2 and Rhodococcus sp. strain Q15 in tandem with YE have an additive impact and therefore more drastically increase the level of degradation of the diesel fuel?
I’m curious to know the differences and similarities between Cycloclasticus and the Rhodococcus species that was investigated in the last paper.
For example, is the transformation and catalysis of a PAH by Cycloclasticus similar to the Rhodococcus process (are the ARHD’s the same, similar, or different)?
Also, I am curious to learn the specificity of catalysis by this bacterial strain. Is growth improved by the addition of YMSM? Are LMW PAH’s or HMW ones more efficiently degraded?
This is particularly interesting because it would insinuate that the marine bacteria have been able to develop pathways able to more effectively degrade PAHs in their specific habitats.
I’m actually surprised that degradation by marine bacteria has not been studied more being that PAH’s stand to cause harm to marine environments. So, I can definitely foresee this study providing unique and useful information regarding not only dangerous PAH’s in aquatic habitats, but also PAH’s in general.
The first thing that really caught my attention when reading this section is that it seems to be significantly more detailed when compared to the first paper. There is obviously more of a genetic focus in this paper, as evidenced by the ubiquitous amount of information relating to the sequencing and cloning. I’m curious as to why that is? Why was it more necessary to delve deeper with this paper?
This section of the paper really reminds me of organic lab, and I guess that’s justifiable. It is interesting to see overlaps between this course and others that I have taken.
One question that I do have, though, is what if some of these transformed products are lost? In organic lab, you may just have a low yield, but here, the products seem greatly more important. For example, what if excessive washing were to remove one of these converted products and therefore that information was entirely left out of final analysis?
I’m also interested in the relationship between the lac promoter and the expression of the dioxygenase genes.
My first thought when I read this section was that there shouldn’t be any associations between these systems because they seem to be unrelated.
I’m still wondering how the particular orientation of the PhnA constructs allow for proper expression under lac promoter control. Perhaps there are overlaps between the regulations of these genes?
It seems that the information provided at the end of this section insinuates the order of the phn genes is significant.
I wonder, since it was not discussed in the past paper, what the order of the phn genes were as it relates to the previous PAH degrader. How big a role does this order play in the ability to degrade PAHs? Is there a specific order that provides a greater degradation efficiency in certain environments?
I’m interested to know the details of why this particular gene possessed high homology with the Rieske center (and whether or not that is even relevant). I wonder whether or not there are other dioxygenases that are quite as effective as this one. Is the Rieske center constant among PAH degraders or are there others that have weaker, or perhaps stronger, impacts on degradation?
I notice that only in this phylogenetic tree is Rhodococci present. It’s interesting that a Rhodococcus does not exhibit the highest homology and there are also other strains that are very distantly homologous.
This really calls attention to the diversity of these dioxygenase genes and the differences between those bacteria that possess them.
I wonder then, since Cycloclasticus seems to be specialized for marine environments and Rhodococcus for terrestrial ones, if there are certain bacteria that are specialized for PAH degradation in other environments not yet explored in these first two papers.
This is interesting to me because it really shows the specificity of these dioxygenase genes. Obviously, there is something very different structurally existing in 3-Methylcatechol that allows for such a high level of activity by the enzyme here.
This then makes me question the evolutionary impacts that may have caused this. Perhaps this particular substrate was more common to come into contact with this enzyme and therefore it developed a higher specificity for it. I may also be looking too far into this…
A study that would look into the specificity of different types of dioxygenase genes would be extremely interesting. Although this study does touch on that, more information on the trends of these genes in different organisms may be able to give a more comprehensive outlook on their functions in different environments, in the presence of different substrates, and therefore their efficiency in different situations. Very enlightening paper.
I would be intrigued to know the evolutionary information behind this type of expression mechanism. Clearly it is extremely complex and very highly organized, so it would definitely be informative to know the role that evolution has played in this system. How fast were adaptations undergone in this system that made for the high level of adeptness we’re reading about? Also, are there potential ways for us to alter the expression of these genes to increase the efficiency of certain strains’ degradation capabilities?
I am curious to know the extent of involvement of the Archaean assemblage that the author(s) allude here. It seems that, as of now, their specific contribution is not quite elaborated upon. Are the Archaeans acting to actually form the biofilm itself? Or are they acting to augment the success of the arsenic metabolic activity? If they are impacting the metabolism, I wonder if this has evolved in this particular environment as a form of symbiosis? And if so, is it obligate or simply beneficial to the Archaean aggregate or the Bacterial bulk? This seems to be important information, but further reading may prove otherwise.
After reading through the paper, I’m wondering whether or not this type of metabolism with arsenic is ubiquitous… I understand that the author(s) refer to arsenic resistance as a “phenomenon”, but perhaps this type of unique metabolism has just not yet been characterized in depth. It makes me feel that certain microorganisms can use almost anything to their advantage if forced to, through evolution. Very amazing. Also, I wonder if there are other avenues through which this type of metabolism can succeed other than that of the biofilm?
One thing that stands out to me from the last paragraph is the differentiation made between the red and green colored springs. Here, it is stated that the samples were mostly taken from the red springs. I’m wondering whether or not this is just because the red springs are less rare, or if it has to do with the temperature…Or maybe they were the only ones with the arsenite traces and anoxic environments… I’m not sure if this is relevant but I’m curious why one was targeted over the other. Maybe those more rare green springs could have provided some interesting information to this study.
Another topic that made me curious about specific collection methods was the use of the “sterile toothbrush” for the slurry creation vs the use of the “sterile spatula” for the DNA analyses. The only thing I can think of here is that it is more important for the DNA sample to be more intact. But that makes me question why the sample taken for the slurry was not required to be fully intact/uniform. This, again, might be irrelevant…
This paragraph specifically mentions electron donor experiments, but never discusses electron acceptor experiments… Why is this? Would it just be redundant? Is all the valuable information discovered in the electron donor experiment?
What is meant by “killed control slurries”? It is the obvious answer that the organisms in the slurries have been… killed? Or is there something else important to recognize? And also, why is the autoclaving, which I am guessing did the “killing”, necessary to perform prior to the radioisotope amendment?
This section in a way relates to my first question in the introduction. I asked what role the Archaea play in this biofilm… It’s not a surprise that there is more diversity in the Archaeal groups because they are considered to be extremophiles. But I’m curious as to the extent of their contribution and the importance of their involvement.
The author(s) mentioning the possibility of an ineffective primer reminds me of the same problem that some of us ran into in our own testing. In this case, because they’re employing PCR, could this be due to the fact that there exists a new and unique gene that has not been previously characterized, as Dr. SNC referenced in the beginning of this section?
When I first read that there was an inability to discover related aoxB, my initial thought was that they must’ve found a novel system… I realized after that the more likely scenario was the primer incompatibility. So, I feel a little better knowing that it’s actual a possibility this mechanism could be something that has not yet to be discovered by anyone. Super interesting!
It’s really amazing to know that some of these extremely intricate metabolism mechanisms are almost as old as the planet. It really puts evolution into perspective… Some things have changed drastically while others have been maintained and stayed relatively the same. Our own evolutionary history is minuscule compared to this!
I think that they had a pretty good idea what was pushing the As(V) reduction, but they still needed to verify their ideas to be able to better understand the entire mechanism. This makes me so curious about how the system evolved and the types of adaptation that it had to go through in order to get to the point it is at today. Super interesting.
One question that I had throughout reading this (I also left some questions, too) was what is the exact role the archaean population playing in this system… It’s interesting that they actually have no involvement here because I expected their to be some kind of syntropy or symbiosis… It seems like they’re kind of just tagging along for their own benefit but they’re not providing anything of substance to the mechanism. Honestly a little surprised by this.
I too thought that the bioaugmentation was interesting. It was said to be the best method over biostimulation in the bioremediation of soils with low indigenous PAH-deegrading bacteria. This method goes against the Antarctic Treaty, which prevents the introduction of foreign organisms into the continent, unless it is used with native microbes. To get a better understanding on how bioaugmentation works, I found a simpler example of the method that used nicotine as the pollutant and synthetic tobacco wastewater as the medium for bioaugmentation. Tobacco is associated with the release of wastewater that contains toxic substances including nicotine. The synthetic tobacco wastewater showed a significant increase in nicotine removal.
This also stood out to me as well and I read that Antartica’s ecosystem is highly sensitive with pollutants and decomposition processes are extremely slow in this region. It’s very great to see a country that protects its environment and citizens from foreign substances.
This was a great point to bring attention to. I read that temperature is definitely a limiting factor in bioremediation. The temperature plays a major role in the rate and degree of microbial hydrocarbon biodegradation. This affects the viscosity of hydrocarbons. So, it is advantageous to increase the temperature when dealing with biodegradation in polar sites and this can be achieved by land farming.
Iron is an essential nutrient for growth and bacteria have to compete for the nutrient. This type of competition id indirect as it describes the rapid culture of a limiting resource. Siderophores come into play because they are used by bacteria to acquire iron. The competition comes because some bacteria are able to produce siderophores, while others are known as “cheaters” and only use siderophores but do not produce them. Species that produce siderophores with the highest affinity for iron has an competitive advantage.
Since V.harveyi cannot produce or import aerobactin, will this vibrio just be deleted from the study. Can it not act as “cheater” as the other vibrios?
After reading the title of this section, I really do not understand. Are they testing for chloramphenicol sensitivity to see if the vibrio is susceptible by the antibiotic or sensitive to it and is this apart of the mutant construction?
In the introduction, it said that aerobactin of V. fisheri prevents the growth of V.harveyi. Is this why the V.fisheri was chosen to become the mutagenesis. If not what is aerobactin?
This image was very important to the study because it set the tone for all other results. Here we see that the ES114 fluid contained an inhibitory substance that allowed for no growth. This is supportive of they hypothesis.
This is very important to understanding how and why V.fischeri produces aerobactin. That’s because it is needed for colonization of the v.fischeri’s host. The relationship between the host and itself is not harmful to either species, so it is more common that these relationships will form. It is just important for them to come together in the ocean that is sometimes depleted of iron.
Nitrification promoting marine nitrogen loss are associated with the OMZs because they have high organic matter and remineralization. Just to be sure the remineralization is the process of nitrogen converting back to ammonium.
So because the Crenarchaeota are autotrophic, meaning they can consume inorganic substances like carbon dioxide, they are more important nitrifiers than abundant AOB?
Great point! I was wondering why Mn was included as they are indeed toxic to nitrification of bacteria. amoA gene would decrease because it is one of the genes encoding for the 1st step in nitrification.
Were the anammox bacteria already determined before it was verified by the CARC-Fish, and if so which step did this happen? I don’t understand how the bacteria was found to bring about nitrogen loss.
Is the Anammox bacteria and proteobacterial AOB abundance the same thing because if they were absorbed at the same depth then is there that much of a significance?
Yes, I feel this is correct Ryne the nutrient availability and oxygen content is the reason for the diversity. More life is able to withstand life here.
So, this “main objective” seems quite vague to me. I mean, I understand what they are saying, but “some naphthalene degrading bacteria” could mean 1 or 2 or all they find. I’d want to know which bacteria they were characterizing and why.
It has always escaped me why scientists work on “should” assumptions. If the gene being studied “should” be critical, then why do we put the cart before the horse here instead of proving or disproving the function and overall dependency of the process of the gene?
How did they come to the conclusion that the bacteria was going to be a good candidate? They say it like it is a forgone conclusion, but that’s a pretty big assumption.
Okay, I get that they added these chemicals to the medium, but what was the purpose of each? I notice that here it says the % of naphthalene degradation was calculated using this formula, but in another place it says it was “estimated.” Were these talking about two different things?
We have the shaker keeping the naphthalene oxygenated, but why would they not consider the effect that moving water or stable soil will have on the growth? This would definitely affect what is there.
So, as far as “biochemical” identification goes, how accurate is this? Of course I can’t name any off hand, but I’m sure that different species of bacteria can have some identical metabolic processes, can they not?
So, I understand that we want the oil degradation, and I consider this a good thing. But, what about the byproducts of the bacteria and their effect on the environment? We don’t want another dead zone like we have in the GOM though.
Just thinking about the big picture here…if the PAH’s are detrimental to mammals, why are we not also concerned with the effect the NDB will have on the Mammals and on the ecosystem as a whole? I mean, did we not just create a miles wide dead spot with the “cleanup” of the BP Oil Spill in the gulf?
Why are we only estimating the amount of naphthalene left? I googled it, and there seems to be several measuring methods for air, but not for water. Does the spectrometry not work for salt water samples?
In the introduction, they say that naphthalene and phenanthrene degrading bacteria have already been well characterized and that here we were looking at more diverse and quick adapting bacteria. Why then are we measuring degradation of phenanthrene here?
This is interesting, as I have read articles related to the effectiveness of bio-remediation using microbes to either absorb or degrade the oil in recent oil pollution. My question is, are there any long-term effects to introducing this organism to this environment?
The certain adapted bacteria inocula may be a key factor to discovering bigger solutions. I am curious to know the structure of the bacteria and how it bonds to each other, ie. does it create a net to degrade the PAHs?
It seems as if the P. Cirtonellolis would be an ideal material to use in bioremediation. It is important to consider the consequences that may arise when using PAH degraders.
Considering pyrene seems to be a difficult compound to degrade, BC1’s ability to do so is interesting. I am curious to know if BC1 is related to reputable PAH degraders.
“Bioemulsifiers act like surfactants but are more efficient in improving solubility…” This is the key to one of my previous questions. This is why BC1 was able to degrade the pyrene so well.
Does the darkness effect the success of the experiments? Also, what is the purpose of using KOH-soaked filter paper? Does the potassium act as a nutrient?
The fnr gene is responsible for converting the selenium, producing red colonies. Its mutation is what disabled the E. cloacae from converting Se(VI) to Se(0). I had trouble understanding this originally.
I am still curious of the significance of the conversion of the Selenium. When thinking of the big picture, I understand we want to identify the genes and pinpoint mutations, but is there a significance to Se?
This concludes that the Enterobacteriaceae would be a potential bacteria for transforming selenium. I would be interested to see additional research to confirm this.
Whats types of environments do naphthalene degrading bacteria thrive in the most and what types force them to compete with other microorganisms the most?
I also agree that Madelyn’s definition of Bio-remediation was very helpful in understanding what this paragraph was trying to say. However, after just reading the articles explanation of it, in did not think it was safe. I did external research to find out that to complete bio-remediation they use natural biomes from soul and groundwater and just add more. My question is how, do they speed up reproduction in the organisms that are already there?
I believe that if bio-remediation does not remove PAH’s then it only does half of the job. Bio-remediation was said to be the most inexpensive way to do this, but it only handles degrading of components of crude oil. My question is what technique is in place to remove PAH’s and their fused rings?
What is the potential for pathogenic activity occurring in the in the marine population? These particular bacteria are pathogenic, how would overgrowth and infestation be controlled?
Studies show that the microorganisms are sensitive to pH changes. If the microorganisms involved in the bioremediation convert the PAHs to CO2 and water, how much CO2 is generated and what is the local effect of that increase on the microbes and or the local marine life?
Is the GC-FID measuring “growth” of the NP degrading bacteria, if so wouldn’t that destroy the bacteria, or is it measuring the naphthalene remaining in the media?
Has there been any research on higher molecular PAHs? Is there a real correlation between naphthalene degradation and their ability to degrade larger molecules?
What is the benefit of the Biolog-GN microplates versus regular plates? Does it allow for a greater number of cultures or does give additional information than other types of plates?
Does transforming E. coli with the DNA of strain ZX4 to E. coli make it easier to observe the properties of the DNA encoding for the meta-cleaveage genes? Is this a common practice to transfer DNA instead of studying the isolated bacteria itself?
Generally the closest genes will be transcribed together while others may or most likely will not. The genes phnH and phnI are only 19 bp apart and would be transcribed together. The phnG gene however, is 181 bp from the first gene and also is noted that there is a spacer and promoter sequences. This would prevent the transcription of phnG occuring with phnH and phnI.
Would the lack of similarity of GST in ZX4 to S. paucimibolis indicate a mutation in the bacteria? Is this related to the partial ORF for GST or is there a correlation between those two?
I believe the HMS (Hydroxymuconic-semialdehyde Hydrolase) is responsible for the step in the catechol, a substrate in the phenanthrene degradation. HMS hydrolases the 2-hydroxymuconic-semialdehyde by adding water and removing formic acid.
If the HSM was not available I would assume the pathway would stall and the degradation would not be completed.
Based on the information in this study as well as others that I have read, Sphingomonas paucimobilis appears to highly effective in the degradation of PAHs. In addition to the PAH degradation, Sphingomonas paucimobilis is also able to degrade xenobiotics.
The dissimilatory Arsenate reduction uses sulfide to reduce arsenate. In another paper I read, 1 mol of Sulfide oxidized to Sulfate would require 4 mols of Arsenate reduced to Arsenite.
Is sulfide the only inorganic anion used in this type of cycle?
Also in the other paper I read, the dissimilatory reaction was done in a curved, gram-negative bacteria also in the Mono Lake, California.
If the Arsenate is reduced to the Arsenite through this pathway, how is the Arsenite oxidized when it says it the biofilm is anaerobic and the photosynthesis is anoxygenic? Also they were no able to obtain PCR data for the Arsenite oxidase gene.
For the bacteria that have both oxidase and transporter genes, how does this affect the microbe’s ability to utilize arsenite or arsenate. I read another article that indicated that there was some resistance inferred if the microbe has both genes.
From what I have read, there is a membrane respiratory arsenite oxidase (aoxB), a membrane respiratory arsenate reductase (arr), and cytoplasmic reductase (arsC). There is also a membrane transporter (arsB).
The arsenite oxidase oxidizes arsenite from the membrane as arsenite is used as electron donor on the cell membrane.
The arsenate reductase reduces the arsenate to arsenite again on the membrane.
If the arsenate is not reduced by the respiratory reductase in the membrane, arsenate may work its way into the cell. The arsenate molecule is very similar to the phosphate molecule and can enter the cell through the pit protein in the cell membrane. When this occurs, the cytoplasmic reductase then reduces arsenate to arsenite. I think arsenate has to be reduced in order for it to be removed from the cell. This is when the transporter comes in and removes the arsenite molecule to prevent cell toxicity with arsenite. Arsenite has a higher toxicity rate than arsenate.
There is a respiratory oxidase(aox) in the cell membrane that oxidizes the nitrite to nitrate on the in the cell membrane.
There is a respiratory reductase (arr)which is in the cell membrane and reduces arsenite to arsenate.
Neither of these processes has the arsenite or arsenate enter the cell.
Arsenate, being very similar to the phosphate molecule, can get into the cell using a phosphate transporter membrane protein. ONce in the cell, the cytoplasmic reductase (arsC) acts on the arsenate and reduces it to arsenite in order for the transporter protein in the membrane to export the arsenite out of the cell. Arsenite is much more toxic than arsenate.
Without the cytoplasmic reductase, the arsenate molecules would accumulate in the cell and kill it.
If I am understanding this correctly, in the anaerobic, dark environment, the arsenate did not reduce easily without out additional electron donors. With sulfide added arsenite was reduced to 1.75 mM. With H2, arsenate was completely reduced.
Does the concentration of sufide a 2mM affect the reduction? If there was a higher concentration of sulfide would it have been able to further reduce the arsenate?
If the bacteria activity was not changed by the addition of oxygen, would that show they are not using water for electrons or oxygen for the electron acceptor?
H2 appears to be the best electron donor in the dark reactions. The redox reactions were much faster with the additions of H2 than with H2S. Incorporation of C14 was higher with H2 as well. Although there is no test done with H2S, I would think the incorporation rate of C14 would be lower than H2 as is the reaction rate.
What would cause a bacteria to use the arrA gene versus ars B gene? Would it be driven by an overwhelming concentration of arsenic?
Or is it triggered if arsenate is able to enter the cell and then the ars C gene reduces it to arsenite and ars B just clears it from the cell? Is this just another reduction process?
Some articles I reviewed indicated that HMW PAHs, that were not degradable or poorly degraded, were successfully degraded, such as fluoranthene, when cultured with phenanthrene.
Is the phn a method for genetic transfer? I was trying to read about the genomic islands and phn and I saw some different GEIs such as PAIs for pathogenicity island but was not certain what the phn is. I thought maybe it’s phn for phenanthrene degradation.
In curiosity, I looked for the composition of the 216L agar. I found this description of this agar in this article: Yuanyuan Fu,1 3 Xixiang Tang,1 3 Qiliang Lai,1 Chunhua Zhang,2 Huanzi Zhong,1 Weiwei Li,1 Yuhui Liu,1 Liang Chen,1,3 Fengqin Sun1 and Zongze Shao. 2011. Flavobacterium beibuense sp. nov., isolated from marine sediment. International Journal of Systematic and Evolutionary Microbiology. y (2011), 61, 205–209.. The agar is composed of: sodium acetate, 1.0 g; tryptone, 10.0 g; yeast extract, 2.0 g; sodium citrate, 0.5 g; NH4NO3, 0.2 g; seawater, 1 l; pH 7.5. What classification of agar would this be? I was thinking enriched?
Is the Cre-lox recombination method a knock-out process? If I understand the process, the Cre recombinase will cut a section of the DNA out, invert or transpose it. The lox is a marker that is on either side of the target.
In comparing the P73T genome with the C. baekdonensis
B30 genome, it is curious that they are similar in genetic material but the functions are different. With the genome similarities would it be possible that the PAH degradation genes are foreign and on plasmids?
Also in another section (paragraph 35) it states that the B30 region was unable to degrade PAHs but in this paragraph states “only 3 RHDs were identified in the B30 region”, if there are 3 RHDs I would think it could degrade something.
I may be just confused but, in paragraph 2 the report is there were numerous hydroxylating dioxygenases and ring cleaving dioxygenases for PAH degradation and then in paragraph 4 it is reported that the C-7,8 dioxygenation pathway is the only one. Did they find potential genes but expression of only one pathway?
This is very interesting and highlights that in nature there is no one reaction. It is interesting to me that they are researching foreign bacteria for the bioremediation. Wouldn’t it not be reasonable to isolate native bacteria?
I would have thought there would be bacteria capable of degrading the PAHs in the area and some that would also have acquired heavy metal tolerance.
Symbiosis at work!! If my impression is correct, the ryegrass is able to degrade, or remove or just hold on to the heavy metals which decrease the efficacy of the degradation enzymes so the bacteria are able to degrade the PAHs.
With the insolubility of PHE in water, methanol is used to dissolve PHE for inoculation in the soil. Then CUCO3 is added to the soil. What is the significance of deionized water?
A change in the natural environment that would reduce the bacterial population thus reducing the degradation activity such as pH changes or moisture. I read some information that chemical oxidation, potassium permanganate, reduces PAH concentration minimally. The decrease PAH concetration is minimal but higher degradation amounts were achieved with a pH 7.5-8.
But I would think those would occur naturally and not in a controlled environment??
Copper is a trace element in plants as well as humans. Copper is involved in defense responses in plants and enhances some plant enzymes. It kind of makes sense that, to an extent, copper would enhance plant activities which in turn assists the bacteria by nutrients needed. I had also read that copper is also directly affecting the dioxygenase activity. Is that correct?
I thought the heavier metals such as iron generally inhibit the degrading enzymes. After reading this through again, I am not sure about the last statement. Other than a toxic level of copper would it replace iron in the enzyme. I thought copper is less reactive.
Is there a relationship between the concentration of PHE and Cu that increases a higher Cu concentration for the plant? Cu does play a role in enzymatic activity but I I can’t find a direct answer that relates increase uptake of copper by the plant. Unless it is just the cycle.?
Yes it is the same process however it is done under nucleotide blast instead of the BlastP that we ran for the RHD genes. The Mega process would be the same except is again is nucleotide not protein.
Could the inability to identify the gene because of non-matching primers? Could there be a missing “catalyst/inducer ” that initiated the reaction or improved the process ?
Is the third degradation pathway very likely to occur? It appears to be a less favorable pathway to me. This reaction would be from a mono-oxygenase instead of the same dioxygenase as the other two pathways?
Is there a benefit to using the phosphate buffer? I read that it the Tris buffer is used during enzymatic processes and nucleic extraction. Phosphate buffers are not affected by change in temperature but Tris is.
Primer walking uses digests of DNA to sequence the genome. A primer starts a strand of DNA and that fragment is sequenced by chain termination(Sanger). Then the next primer used is formed from the end of the sequenced strand before. The process would continue until the genome is sequenced. If I am understanding this correctly.
Why is flnA1-flnA2 not able to degrade Carbazole? If I understand this correctly, Carbazole has a hydroxyl added then flnA1-flnA2 acts on it and becomes a dihydrodiol and flnA1-flnA2 changes to a linear action instead angular??
The flnA1-flnA2 dioxygenase complex is able to produce the three oxidation products listed in Table 2. DFDO is able to only produce 1-Hydro-1,1a dihydroxy-9-fluorenoned and CARDO is only able to produce Dihydroxyfluorene with Fluorene as the substrate and neither of these produced Fluorenol-dihydrodiol. Is this a specificity or approach different in the three dioxygenases?
I think what they are saying is that the majority of studies with angular dioxygenases have been in gram positive bacteria and the primers tested were from gram positive bacteria. Perhaps that contributed to the lack of amplification.
The granule version of Carbofuran was in fact banned in 1994. It was banned for bird deaths from ingesting the granules as well as the deaths of animals that ate the birds contaminated with the pesticide
This paper is intriguing. I appreciate the difference in the subject matter, meaning this is an intentional act to “protect” crops that may actually be causing greater consequences.
I am having some confusion in this paragraph. Is the identification of the metabolites of Carbofuran identified by comparing to known pathway metabolites using the processes listed?
In this situation, these are bacterial conjugations used to randomly transfer the Tn5 transposase gene and Km resistance to construct a mutant library.
Would the loss of the expression of the gene cluster cfdABCDEFGH be because the CftA gene is a transporter and without the transportation there is no need for oxygenase expression?
In reading the Fig 3, It looks to me as though all the mutants as well as KN65.2 can complete the initial step,as the concentration of Carbofuran decreased from 0.8 to 0 either immediately or within 2 days.
In this paragraph, the mutants in group V are not able to degrade the carbofuran completely thus accumulated carbofuran phenol. The mutation of cftA not only interrupted the ability of carbofuran into the cell but also decreased the expression of the gene cluster believed to degrade carbofuran. As the accumulation of the metabolite 1, carbofuran phenol, increased, an alternate transporter gene was strongly increased to remove the toxic build up. Am I interpreting this correctly?
Would this expression similar to lac operon minimal expression? Or could it be expressed for a different purpose? Aren’t some of these genes involved in some type of fatty acid synthesis? (Making enzymes that aid in the degradation).
Hey Julie,
I too am fascinated by Rhodococcus and its ability to transform harmful PAHs into a TCA cycle.I believe it has a promising future for the clean up of polluted areas. Also as you’ve listed in paragraph 2, “PAHs are found in coal, wood, and other materials.” The Rhodococcus strains are in fact efficient at removing hazardous elements such as sulfur from coal and petroleum that are found in PAHs, thus benefiting our well-being.
I thought that Rhodococcus belongs to the family of enzymes known as ARHDs which should have exhibit homolgy with catalytic domain even for previous species? Could there been a change in its bioremediation? Did bacterial evolution change its functions over time?
The thin layer chromatography was a fun experiment somewhat similar to streaking in my opinion. Analyzing the spread of both and the migration of the spread over time, also being careful not to rip through the surface while doing these experiments.
Could colonies grow even after a certain period of time? Also Does MSM and R2A plates form different numbers of colonies or does it depend on how well you perform the experiments?
Knowing what I know now about MSM and R2A plates, Im definitely getting more comfortable with the lab preparations such as being able to streak and being able to describe the colonies just by looking at them. For example the different colors blue or pink, characteristics in the colonies such as pure culture and observing the growth of them with time.
The R. erythropolis in YMSM in figure 2 shows on day 1 to 3 the residual Fla percentage dramatically dropped about 60 percent while the R. erythropolis in MSM shows a slight drop. Gradually over time the MSM also had a huge drop in percentage from day 5-7. As for YMSM there was just a constant percentage drop.
hey anna , I also think its really cool and fascinating that Rhodococcus species come from a diverse tree with mycobacterium being the best relevant match. Also the tools to figure out the closely related, its specific gene, and the number of nucleotides. Maybe its possible that they have the same functions because they do seem to have very similar attributes.
I read more great things about Rhodococcus with its unique functions and diverse abilities that make them such a special case. Im glad that there are studies in pharmaceuticals and chemical industries that will help benefit the environment. Im hoping that Rhodococcus will become a well known compound that performs efficiently in many diverse roles in the near future.
How efficient is the tool to amplify “bp” measurements? Also in this case does Rhodoccus sp.CMGCZ expected to not go through with its regular degradation with a different bacterial strain PAH dioxgenase gene due to bp? How close does the bp band have to be for regular degradation to happen?
I find it fascinating that Cycloclasticus has been reported as another bacteria that is able to degrade PAHs in marine environments. I was in belief that Rhodococcus was only the bacteria to degrade PAHs, I’m interested to see the breakdown of this bacteria and to see if there is any similar functions or characteristics as Rhodococcus.
Im wondering what the level of toxicity is for PAHs? For example when dealing with burning of grass fires or even smoking cigarettes, do you immediately react to the exposure of PAHs or is it long term effect? Also now there is a possibility that seafood consumption is contaminated. Hopefully Cycloclasticus is capable of clearing the contamination in marine environments.
Im wondering if sonication was purposely used in the experiment. Was this effect used to open the cell for further use and the result was supernatant as the crude cell extract?
It’s interesting that phnA(1-4), phnC, and phnD genes are key players for the degradation of PAHs. I notice that the organism Sphingomans sp. exhibited the highest percentage of identity labeled on Table.3, also showed the strains had the best sequence with the amino acid of ISP subunits of aromatic oxygenesases. Im wondering if these genes are effective to the degradation of PAHs.
Ive noticed that phnC reveal highly significant percentage then any other gene. It’s possible that phnC has the most potential for example, high activity with different substrates. Im interested to see how well does phnC contribute to the Cycloclasticus in degrading PAH.
After reading paper 2, I believe the phnC gene is the best candidate for bioremediation because it exhibits 60-66 % identity with amino acid sequence(highest group) and can degrade PAH dioxygenase efficiently.
Its interesting that PhnC gene encodes the PAH extradiol dioxygenase with substituted monocyclic catechol compounds. Also its involvment of both upper and lower pathways for degradation of napthalene, phenanthrene, and biphenyl. Im hoping researcher will find the true substrate of PhnC that gives them a better insight on the pathways of degradation in Cycloclasticus.
Im surprised that some microorganisms are able to withstand normal existence when high concentrations of As(III) and As(V) are present. Im excited to see the results in oxic and anoxic, however I believe researchers are focusing arsenic under anoxic conditions because it’s only dependent on the amount of organic matter. It may be predominantly better assuming the results will be more efficient. -I may be wrong
It’s interesting that the light driven from anaerobic ecosystem may have played in the Archean Earth. It’s possible that this anoxic environment used light such as redox couples As(III) and As(V) serving as electron donor and acceptor for obtaining energy to survive and later evolved.
Its interesting that slurry mixture were stored and preserved in the dark for several months prior to the experiment. My question is whats the reason for a mixture to be left for so long?
The light and dark incubation having an effect on the electron acceptor and donor of an arsenic. Does this change the integrity and activity of a bacterium? For example the photosynthetic bacterium Ectothiorhodospira strain PHS-1, can it evolve quicker under light with As(III) condition vice versa of a different situation using dark.
In order for bacteria to thrive in harsh environments I would assume that they would have similar structures to Archaea since Archaea are known to live in harsh environments.
I would assume that the explanation dealing with the primers being not suited for the environment and the solution being to try a new primer is more plausible than it being a new microbe.
It’s interesting that the microbial populations were more effective in light incubation expressing similar pure culture.
Im wondering if dark incubations had a specific temperature will it change the process of biofilms.
Its interesting to see that As(V) reduction could be viewed as an ancient phenomenon that dates back 3.8 billion years ago. I actually would have loved to see more about the evolution and phylogenetic diversity.
Since the structure of PAHs contributes to their biodegradation resistance, how does degradation take place in PAHs? Does the fused ring break apart first?
Anna, I am also curious as to why the phylogenetic trees were constructed. Were the phylogenies used to determine which bacteria was a more efficient PAH degrader? Or did the phylogenetic tress help the researcher have a better understanding of how each bacteria played a role in degradation?
Why was the temperature taken at a depth of only 1cm? Would taking the temperature deeper into the water yield different results? I am curious about how the temperature of the soil sample affects PAH degraders.
In locations B and C, the concentration of extractable PAHs and the ionic concentrations are high. Locations B and C had higher ionic concentrations than location A. Since these locations are rich in nutrients required for bioremediation, would there be a higher success of natural bioremediation in locations B and C than location A? Or would the success rate be nearly the same due to low nitrogen and phosphorus levels and the high PAH concentration?
Since nutrient-rich media favor rod bacteria over coccoid bacteria, would we only see rod bacteria in the different morphologies? The text stated that rod bacteria can out-compete cocci bacteria due to their surface contact. If both bacteria are together, would rod bacteria take over completely? Or would a small amount of cocci bacteria be present with the rod bacteria?
Anna, I struggled to understand this section, but after reading your comment about using this data to construct the phylogenetic tree, this makes sense. I think that creating the phylogenetic tree helped in the identification process. This section outlines the similarities that the isolates have to classes under the Proteobacteria phylum. The next paragraph explains the similarities in the isolates and the classes, as well as degradation processes within the classes. I agree, the construction of the phylogenetic tree does lead you to think that related bacteria can be useful PAH degraders.
The text states that there is almost no record of the use of P. citronellolis in PAH biodegradation. Since it is not specified as a pathogen, and it had an astounding amount of growth on the PAH substrate, why haven’t more experiments been performed with P. citronellolis? It seems as if P. citronellolis would be very beneficial in PAH biodegradation.
I am a little confused on how the hydrocarbons are recycled. Do the vent fluids bring the petroleum hydrocarbons upward from the deep sediments? Or are the petrochemicals within the vent fluids responsible for the hydrocarbon recycling?
The text Staes that not much has been published in regards to deep water hydrocarbon degradation. Could hydrocarbon-degrading bacteria present in coastal waters also be beneficial in deep waters? Could the same bacteria perform degradation in deeper waters, or does the depth and temperature of the water play a role?
I am also interested to see if each sample contains unique PAH degrading bacteria. Core 4567 was collected from a deep water environment, with no sulfide, and no bacterial mat. Core 4571 was collected from a deep water site, but it was oil-abundant with a bacterial mat present. Core 4571 also had sulfide present. Since each cores were collected from sites with very different conditions, I would expect the PAH degrading bacteria to have different characteristics regarding to where it was collected.
I am unsure of how the aluminum foil prevents hydrocarbon sorption. Does it serve as an extra protective barrier around the cap? Another thing that stuck out to me was that all of the flasks were incubated on the orbital shaker in the dark. Is there a reason they were incubated in the dark?
By looking at the graph provided, it seems as if there are drastic differences in the amount of 14CO2 recovery from each sample. Compared to core 4571-2 (solid bar), core 4567-24 (open bar) seemed to have a great amount of 14CO2 recovered for NAP,PHE, ANT, FLU, and PYR. Core 4571-2 (solid bar) had a higher amount of 14CO2 recovered for BaA, but it was not as drastic as the others.
The study found that Cycloclasticus could be a major hydrocarbon degrader at the Guaymas Basin. If Cycloclasticus co-exsists with other hydrocarbon degraders in surface sediments, what makes it have poor cultivability and hard to isolate compared to the other hydrocarbon degraders?
Performing microbial bioremediation is particularly difficult in Antarctica due to the extreme climate and constant low temperatures. Microbial bioremediation processes are slower, compared to the rate if administered in warmer or tropical regions. It also affects hydrophobic contaminants in the region such as the PAH from diesel oil. Hydrophobic (lipophilic) contaminants tend to be more durable and more difficult to remediate in low temperature regions due to the slower biodegradation rate.
– I was not familiar with the term anthropogenic pollutants or bioaugmentation as a remediation strategy for pollutants. An anthropogenic pollutant means that the source of a pollutant is a direct result of human activity. Bioaugmentation is a microbial bioremediation process that works by adding a specifically isolated hydrocarbon-degrading bacteria that have been cultured in the lab (ex-situ). Indigenous Antarctic hydrocarbon-degrading microorganisms are often the best adapted to the environmental conditions. For this reason, they are a good candidate for bioremediation/contaminant cleanup from oil spills.
The multi-process technique for phenanthrene quantification caught my attention, and I did some further research. It seems that every variable in this study is substantially complex and highly specific. In this case for quantifying three-ring PAH concentrations, the combination of excitation-emission fluorescence spectroscopy and multivariate data analysis are useful for modeling degradation studies and monitoring PAH concentrations and microbial growth under Arctic conditions. Recent technological development now utilizes multi-dimensional EEMs, derived from EEM fluorescence spectroscopy, scan excitation and emission ranges simultaneously, reducing broad spectra, and increasing selectivity.
The aromatic ring structures within the PAHs and their hydroxylated metabolites possess naturally fluorescing properties. Fluorescence spectroscopy may be used to analyze the concentrations of these compounds to determine degradation and metabolite production. With multiway data and PARAFAC analysis it is shown that reliable concentration determinations can be achieved with minimal standards in spite of the large convoluting fluorescence background signal. Thus, rapid fieldable EEM analyses may prove to be a good screening method for tracking pollutants and prioritizing sampling and analysis.
Microbial remediation is not solely effective in cold regions, but is the most suited technique for harsh, arctic conditions. In cold environments, oil-based contaminants remain longer than in temperate regions due to the low bioavailability of hydrocarbons and the harsh climate.
Natural remediation by Antarctic soil microorganisms is slow due to low availability of liquid water, lack of nutrients to support microbial growth and low temperatures that reduce bacterial metabolic rates. One particular bioremediation strategy suited for cold regions operated by introducing native Antarctic hydrocarbon-degrading microorganisms, as such organisms are often the best adapted to the specific environmental conditions.
Oil spills generally have a negative impact on soil biodiversity, in which overall microbial biodiversity may decline.
Following an oil spill, hydrocarbon-degrading bacteria can increase significantly in number, and may become a major proportion of the total culturable microbial population. This study confirmed that diesel exposure has favored the development of microorganisms that use hydrocarbons as a source of energy. Antarctica’s microbial communities inhabiting regions near an oil spill are excessively exposed to hydrocarbons, which then caused a selection for microbes that are capable of utilizing PAH as carbon and energy source.
Bacterial staining and dye techniques are fascinating and gives me an appreciation for how far science and microbiology has come. The importance of bacterial detection on agar plates is paramount and will indubiously continue to evolve as microbiological techniques advance.
I agree with you, Julie. I believe that the exploitation of rhodococci and their unique ability to transform PAHs into different intermediates can greatly benefit human society and health, as well as the atmosphere. The use of rhodococci could significantly lower the risks of human diseases by removing hazardous environmental toxins.
I predict so as soon as scientists can hypothesize a method to harness the abilities of non sporulating aerobic rhodococci. It is important that more studies such as this are done in order to prevent further pollution and toxic compounds from entering into the atmosphere.
I am interested in this as well. I wonder if there would be cases of negative results during microbial manipulation due to the addition of organic/inorganic substances. I suppose it would all depend on the microorganism isolated.
I was thinking the same when I read about the use of the minimal salt medium. Since it seems to be a commonly used practice in laboratories, I am interested to see how effective the MSM is at isolating pure cultures of bacterial strains for our labs.
I am finding many similarities between the scientists’ methods and procedures and the ones we have used in our lab 2. The spreading of diluted cultures on agar plates seems to have yielded pure cultures of bacteria. I am excited to see how our bacterial children grow and develop after being incubated.
It appears that the bacterial strains had a high affinity for Fla compared to Nap and Phe. The difference in percentages of the PAHs degraded varied greatly between Nap/Phe and Fla. I wonder why in the presence of YE that the strains were not capable of degrading Nap, yet Phe and Fla were still able to be degraded.
I wondered this as well and what made the bacterial strains have such an affinity for Fla compared to Nap and Phe. It probably has to do with the culture mediums on the agar plates, but I am still curious whether there is a way to better select and isolate a substance for degradation.
I am curious to know what kind of pharmaceutical and biotechnological advances we will be able to make with Rhodococcus. Manipulating and exploiting their metabolic abilities will continue to progress modern technology. The use of their enzymatic properties will perhaps give us more insight into the nature of bacterial strains.
Rhodoccocus preferred Fla as a source of carbon over Nap or Phe given that it degraded almost 100% of the Fla and small percentages of Nap and Phe. I wondered why this was after reading the results last time but understand now that it was due to the enrichment and subculturing techniques these scientists used.
I assume that obligate marine bacteria are more capable of degrading PAHs than terrestrial bacterial strains due to the presence of excess pollutants in seawater.
I am interested to see how the Cycloclasticus strain of bacteria compares to the previous PAH degrading Rhodoccus bacteria in the last study. It seems that it will have more of a capability to degrade oils than did Rhodococcus.
I am interested to know more about how the lac operon was used in this experiment and how it affected the results. With the ability of the lac operon to repress or activate cellular activity, I suppose there are numerous outcomes during experimentation depending on what genes are manipulated.
This interests me because I began writing the pre lab for Wednesday and I recall some of these procedures in the protocol. I am excited to be able to use these techniques on our microbes. Centrifugation is fascinating in that it separates cells into layers due to high speed spinning so that you can visually see the different components.
This passage interests me even more after it was explained in class Wednesday. I feel I have a better understanding in these methods as far as using plasmids as vectors.
The ability of these strains to turn the indole to indigo is very similar to the microbes we are working with in lab. This makes me curious about whether our bacteria contains aromatic oxygenase genes.
I, too, find it incredible that we are able to work with genetic sequencing to determine the chemical abilities of microbes. I am also interested in the Cystoclasticus strain and its PAH degradation abilities compared to the strains we have been using in lab.
It is interesting that the PAH degradation genes were located on plasmids in Pseudomonas strains but on a chromosome in the Cyclocasticus species, yet both genes function similarly despite being on different genetic locations.
I am interested to know if further studies will be made involving the efficiency of the other dioxygenase genes of Cycloclasticus. Since it has been established that two different species of bacterial strains that can degrade PAHs have been discovered, I assume there must be many more bacterial processes that we can use to our advantage in industry.
I was also wondering this as I was unaware that it was possible that a bacterial strain was unable to produce a usable PCR product, as this possibility has not been mentioned. I wonder if it has something to do with the activity of the primer or polymerase.
I am actually surprised that bacteria can exist in environments rich in arsenic, arsenate and arsenite considering the level of chemical extremity. I was under the impression that archaea would have been the only dominating prokaryotic organism in these environs.
I find it fascinating that this study will be involving Archaea, as it is incredibly interesting that these microoganisms can live in such extreme environs such as hot springs and regions high in volcanic gases. I am excited to see what these scientists uncover as the experiment progresses.
I am curious to know why the samples were stored in the dark and why this was important to the experiment. I was unaware that that would have an affect on isolates.
I am interested to know why the dark incubated control groups were wrapped with aluminum foil and what the foil’s function is for the purpose of this experiment. Is this just to shield the sample from the light or is there another purpose?
It is incredibly interesting that similar strains from the same red biofilm had vastly different temperature ranges, yet their optimal growth temperature was comparable. This makes me wonder about the differences in their metabolic functions.
I wonder if the use of different kinds of primers suitable for this particular environment would have resulted in better results in regards to the detection of arsenate oxidase genes.
What I gathered from this paragraph is that coastal spills are the most detrimental pollutants to the environment, but there are safe ways we could improve and handle this. I was still a little confused by the concept of bioremediation so I googled the definition which made it much more clear so I could move on to the next paragraph. For those as confused as me… “Bioremediation: process used to treat contaminated media by altering environmental conditions to stimulate growth of microorganisms and degrade the target pollutants”
So if I read this one correctly, is it now saying that bioremediation is not efficient at removing PAH’s which is what they are about to try to do? My next thought is, there has to be a something that can break down the fused rings of the PAHs to make them less resistant.
So now there is reported success with bioremediation used to degrade PAHs, however the trials include co-substrates that help make it successful. They are testing it with no added substrates to see how much the co-substrates were taking credit for in degradation. From the 4 paragraphs i’ve read, my first thought is that the co-substrates helped a lot.
It says that the temperature was taken at a depth of 1cm. Was there any way that the water affected the thermometers readings considering 1cm is not that deep? Is that not just the same as taking the waters temperature? I am confused what taking the temperature was actually for.
Does suspending the pellets in BH medium make a concentrated bacterial solution because the pellets dissolve or because they just contaminate the BH medium? I do not see anything else mentioned about the pellets so I am assuming they dissolved and formed the later BH liquid?
Not a question. Just pointing out that an increase in microbial population results in a clearer culture solution. This stood out to me because I though the opposite would happen until it said that the weight of the bacterial cells makes them settle to the bottom, which then made sense.
So I thought gram-negative bacteria would appear pink in color. Is this only on certain agars? It says these colonies were cream and clear but they are gram-negative. Maybe I am getting something confused?
I found it interesting as well that the rod shaped bacteria can out-compete cocci. It makes sense that they have better contact with the surface, it just surprised me that they were that much more dominant.
No questions. I just took away that natural bioremediation has many studies in deep petroleum reservoirs and coastal environments but very few in deep-sea waters. Since more exploration is happening for oil in the deep-sea, it makes since to learn more about natural bacteria that could degrade any oil mess made.
It seems like deep-sea degrading bacteria live at high temps. This makes me think that they are a different strand or type of bacteria than the shallower coastal water degraders.
I don’t think the -80C will kill the bacteria. We are storing our bacteria in lab this week at that temperature to make the freeze stock of it. Based on this saying “prior to storage”, I imagine they are doing something similar to us storing ours.
So they inoculated vials with the wet sample that was collected… but it says they prepared the killed controls prior to inoculation. What killed controls?Also, it says all treatments were done in triplicate… Is that saying that there were three samples of each of PHE, ANT, PYR, FLU, NAP, and BaA?
Okay so i am confused on how many flasks there are… is it saying they had 15 flasks containing 18 ml of ONR7a medium and 2 g of sediment slurry from core 4567-24 and then they took 5 of them and added 1 mg of [U-13C]-PHE and another 5 and added 1 mg of unlabeled PHE??? I hope this makes sense because I am confused on what they are replicating and duplicating
PCR makes many copies of DNA whereas qPCR targets a DNA molecule during PCR. So i’m guessing they used qPCR since they want to know the amount of the specific OTUs?
They were successful at identifying PAH degraders in the deep-sea in the Guaymas Basin sediments. So what is next? If these PAH degraders exist, would they not already be showing a significant influence on the present crude oil?
Am I reading this correctly… Are they saying they are trying to see if the same Cycloclasticus PAH degrader found in shallow water sediment is also found in deep-sea sediment along with other PAH degraders?
I feel like this paragraph can easily be shorted down for the summary. The main take away seems to be that PHE showed the highest mineralization so they used it to continue on in the experiment.
Im guessing that the main take away here is just the process of tracking C13 with the use of C12 and C14. I wouldnt think knowing that it took 11 days matters for a summary.
So toxic states of selenium (selenate & selenite) can be reduced to selenium with the help of microorganisms. Bacteria that can respire Se use selenate and selenite as electron acceptors and then emit selenium. Even those these bacteria are very common, we still know little about what they are capable of.
1.) The cells were grown in Pyrex bottles in LB Broth 2.) Cell density and oxygen concentration in the pyrex bottles was monitored3.) Cell were harvested from the pyrex bottles and suspended in salt medium4.)Antibiotics were added to the cells in the salt mediumAt what point were the strains grown on LB agar?
The first 5 sentences seem very detailed… is our main takeaway from this paragraph suppose to be that three strains were tested for selenate reductase activity based on the color change on the agar plates?
So the nucleotide sequence of 9.75-kb DNA fragment was determined… it had 8 ORFs… it was then subcloned into 4 fragments…then what is it saying? They looked at these 4 fragments to see if complete ORFs had fnr or ogt genes so they would know which one was responsible for selenate reductase?
So they all came to the conclusion that E. cloacae cant use Se(IV) as its only electron acceptor for anaerobic growth? They are comparing the current experiment with previous studies, right?
So FNR regulation is active without oxygen present… so when there is no oxygen the fnr gene is expressed and that activates Se(VI) reduction. Is that correct?
So here they are saying E. coli doesn’t have the oxygen sensing transcription factors so it doesn’t know if oxygen is present or absent, therefore it cant reduce Se(VI) because the reduction of Se(VI) happens due to FNR regulation that starts in the absence of oxygen… right?
Okay so E. coli does reduce Se(VI), but it is not from FNR regulation activating it. Rather it is from the presence of the YgfK, YgfM, and YgfN proteins. So is this part just saying there are multiple ways to reduce Se(VI)? This discussion section is confusing me.
I am curious why we haven’t seen this as frequently in marine bacteria- is it because they are less studied in general or they have been studied but this mechanism is still not seen? This would be an interesting topic to study among marine bacteria as they could target the marine pollutants as well.
Given the differing environments, I also think this could be a semi-universal gene/genomic island but it is only expressed in certain environments or in differing conditions. It also makes me wonder about post translational modifications or epigenetic factors that could affect the protein expression.
I am looking forward to their methods and results because I wonder if this wide range of degradation is due to one extensive metabolic degradation pathway or several unique pathways. Given that they are looking into the first fluoranthene degrading bacterium, I would think it is a specific pathway but I would not be surprised to see familiar intermediates and mechanisms. In breaking down the fluoranthene, are there harmful byproducts or anything the bacterium can use for its own benefit?
I wonder why they had to predict the remaining protein coding sequences? Were their samples degraded or not fully manageable due to their environment? Depending on the accuracy of this technology, this could affect their results greatly.
I am most familiar with vectors inserting a novel gene to a plasmid, but it appears this vector carries ampicillin resistance and B. subtilis synthase subunit. I am not sure of the significance of the latter but it appears this is also introducing the deletion of the target, knockout gene. I am looking forward to their results and discussion so that this will all start to come together more.
This is very interesting that the P73 strain was able to acquire the PAH degradation properties from a bacteria in another order via horizontal gene transfer!
It is interesting that the most HTG genes were related to the rhizobiales but the greatest overlap in the proteome was directed toward an unrelated strain, C. baekdonensis.
This is really useful results of the study! By narrowing this down, they are able to determine which other genes could be necessary for PAH degradation.
I agree, this is a great, defining statement reporting their unique results. It is incredible that they found a protein that is a part of the most difficult step of fluoranthene degradation. It’s interesting that this is the first toulene/biphenyl protein to be found but it is also able to catalyze other similar materials whose pathways have been previously described.
I hate reading about how we as people are really destroying our planet. Antarctica has been brought up plenty of times in my previous Biology class about how bad it gets contaminated with oil and such.
I am glad I am learning some of the vocabulary in our lecture class. At first I was reading this so confused, but then what a chemotaxis was and how it is increasing or decreasing concentration of a particular substance. Now I understand a little better now.
I like how this paper concluded, it is usually hard to understand some of the material and research being put forth. However, this paragraph helped me understand the overall point being made.
I can see the obvious advantage bioremediation has over physicochemical treatment. Clearly any time we are able to avoid adding more toxic chemicals to the environment we should. However, I am curious about the disadvantages of bioremediation. Will the addition of these microorganisms disturb the balance in the ecosystem? Will other organisms be negatively affected due to increased competition for resources? What happens to the microorganisms after they have fulfilled their purpose of degrading the oil? Will they simply die off? Is this method costly?
If the oil entered and polluted this body of water nearly thirty years ago, surely it has spread and is affecting an even larger area by now. Is it even possible for bioremediation to make a significant impact on an area this large? How long would it take to see progress?
Why were the seawater samples transported on ice? What is the advantage of doing this? I would have thought that it would be best to maintain the samples’ original temperature in order to keep the bacteria alive.
What was the purpose of filtering the seawater and then adding the filter paper to the nutrient medium as well? Couldn’t you just skip the filtering step and have the same result?
The article says that eighteen strains had an adequate growth rate for further testing. However, they only sequenced the 16S rRNA gene for twelve of these. Why weren’t the other six sequenced as well?
If I understand correctly, BATH % indicates a strain’s ability to attach itself to the naphthalene; and it is this attachment that facilitates emulsification. I see that for most strains, their BATH and emulsification values are fairly similar, except for strain N4. Strain N4 has a BATH % of only 5.71, but its emulsification activity is still 62.23. What other methods could this strain be using to degrade naphthalene without direct attachment?
Since the most important gene in PAH degradation is found in a plasmid, how common is it for non-PAH degrading strains to acquire this capability through horizontal gene transfer?
The Bushnell-Haas medium is considered a defined medium because its exact composition is known.
Also, I know that defined media which contain only a single source of carbon are called “simple” defined media. Is there a specific term for media that exclude carbon all together (such as BH)?
From what I understand, this experiment tells us which carbon source best supports the strain’s growth. This is determined by observing the turbidity of each microplate (which each contain different carbon sources). Higher turbidity means more cell growth. More cell growth means more degradation of that specific carbon source. Correct?
This paragraph states that strain ZX4 could be used as an engineering bacterium for producing indigo. Does this mean that these bacteria could produce indigo dye on a commercial scale and eliminate the need for extracting indigo from plants?
So is GST function basically similar to that of catalase, (which removes hydrogen peroxide wastes), except the GST removes phenanthrene components/wastes?
The specific line I want to focus on is “this results in a significant decrease in species richness and evenness, and a large decline in soil biodiversity of contaminated soils.”
Immediately after reading this, my mind shifted to think of a place we are all familiar with–the Mobile River Basin. The largest inland delta system within the United States is filled with an immense amount of species richness. I must say, many of us are not as familiar with Antarctica as we are with Mobile, Alabama. This led me to look into factors affecting our home town in hopes of relating it back to the study. Firstly, the Mobile Bay has a high contamination of water which leads to dead zones. These dead zones have such a high level of nutrients that the cyanobacteria becomes harmful to the region. When looking into Antarctica, the dead zones are claimed to be from the continuous burning of fossil fuels (which is also outlined in this paper). It is fascinating to see that these dead zones spread across all of the oceans across the globe.
The final thing I want to address in this paragraph is the source of these contaminates. Specifically, the addition of paper mills along the Mobile basin. This past year I had to opportunity to work for a paper chemical company directly in the paper mills. This personal connection intrigued further research. Many plants use a bacterial mixture in their waste water treatment before releasing back into the adjacent river. It turns out that this antimicrobial concoction may have a negative impact on animal and human populations. To the best of my knowledge, there are no paper mills in Antarctica, but it does not change the fact that mankind is the source behind the fossil fuel use in the region.
[by directly seeding contaminated sites with pollutant-degrading bacteria (bioaugmentation). Since the Antarctic Treaty impedes the introduction of foreign organisms into the Antarctic continent, bioaugmentation can only be implemented by the use of native microbes]
Since being in this class, I have solely focused on the idea of hydrocarbon degrading bacteria after being in lab. After doing some intense googling about my previous comment referring to dead zones, I happened to come across a nitrogen degrading bacteria. Many petroleums are able to be degraded using hydro-carbon degrading bacteria as listed in this paper. SAR11 is a nitrogen degrading bacteria that has been discovered to aid in denitrification. These bacteria consume nitrate and convert it into nitrite, in turn becoming gaseous nitrogen. This nitrogen then leaves the ocean releasing greenhouse gases into the atmosphere. The enzyme nitrogen reductase is a genome that potentially allows bacteria to breathe nitrate instead of oxygen. In fact, they have tested this on E. Coli and the DNA did produce the enzyme. Apparently, SAR11 is one of the most abundant bacterias in the world’s oceans. With that being said, there is a high possibility this may be a native microbe to Antarctica.
This is a very surface research level on the nitrogen degrading topic, but I am curious in how this compares to the carbon degrading bacteria. This would be a fascinating conversation to have with others to bounce ideas off of.
I thought about this particular oil spill as well. I thought it was interesting because everyone in the States heard about the disasters this entailed, but it wasn’t until a few days ago that I learned about the diesel oil problems in Antarctica. Specifically with bioaugmentation, the temperature of the waters in both regions are vastly different. Since typically microbes respond better to warmer climates, would there be a slightly different process based on where bioaugmentation is used?
[Samples were taken from the surface soil horizon (0–10 cm) from four sites exposed to diesel fuel]
What would happen if a deeper depth of the soil was analyzed? It states that they are taken between 0-10cm, but the severeness of diesel fuel exposure may be different at each sample site. At the different sites, is there a different maximum depth for alteration, perhaps maybe 15cm? I think it would be interesting to look at the results if samples were taken deeper than 10cm to see the effect of the naphthalene degrading bacteria.
Another thought on the soil depth–were the samples taken 0cm, 2,cm, 4cm, etc.? This seems like a way to see the difference in the saturation of diesel oil.
Polycyclic aromatic hydrocarbons are nonpolar compounds with delocalized electrons. My thought process is the idea of like attracts like. The water is polar causing the extremely polar oil to stand out. It seems that the bacteria may have to target nonpolar compounds. This is something I attempted to look up, but did not get a clear cut answer.
When doing some research for this topic, a new question arose. In colder waters, there is a higher amount of nutrients present due to the deep water rising to the surface, hence more phytoplankton present. Does the higher level on nutrients in the Antarctic water have a role in the non-polarity of the PAH?
The paper states “The optimal growth temperature was determined by a previously published protocol (Gallardo et al., 2014).” I looked up the study and it brought me to a writing called “Simultaneous effect of temperature and irradiance on growth and okadaic acid production from the marine dinoflagellate Prorocentrum belizeanum.” It states that this study was run at 18, 25, and 28 degrees Celsius. This agrees with the fact that the M&M we are reading was grown at 28 degrees Celsius.
[Bacterial growth in soil from non-contaminated sites was promoted when glucose was added, but not in control conditions or when phenanthrene was present]
When looking at the figures that are provided in the unexposed site, it can be seen that glucose did indeed encourage bacterial growth. It did start to plateau as the time (in hours) increased but I am assuming that is due to the decreasing nutrients presence as growth continued. This trend line is very apparent. In fact, right at the 72 hour mark, phenanthrene suddenly has a positive growth so much that the statistical bars do not over lap with the non-growing control group. When doing research on the metabolism of PAH, it was fascinating to see that this is how carcinogens initiate lung cancer. There must be an activation by xenobiotic-metabolizing enzymes for this carcinogen pathway to begin. The whole process of metabolizing PAHs is interesting when seeing the potential negative impacts.
It is interesting because Figure1D, shows that E43FB has the second highest phenanthrene degradation level. It has solid growth in the liquid M9 medium with phenanthrene but terrible growth with the diesel fuel (comparable to E.coli). However, the optimal growth temperature was determined to be 28 degrees Celsius. The average temperature in Antarctica is -49 degrees Celsius. E43FB had the most extreme cold temperature growth around the 4 degrees Celsius range. This leads me to your question: if I could predict, I wonder if we would see a change in the strains and they all flip growth rates since E43FB favors the cold temperature. It would be neat to even look at it at its ideal temperature and compare the growth there.
After looking in to xenobiotics too, it was interesting to see that these foreign materials would be extremely dangerous if it was not for metabolism. The PAH degrading bacteria helps to return the environment to the base condition. There are multiple enzymes that must be used in order for them to be properly processed. In fact, in the phase I biotranformation (in the CYPs) is where the metabolism of hydrocarbons takes place. The molecules present in phase II are generally less toxic and go through a conjugation reaction. Finally, there is a phase III but from my understanding the research is limited on this topic. It seems like this is the best place to explore and see the potential for PAHs and their enzyme-inducing properties. Since we can only have native substances in Antarctica, where are some other places we can pull enzymes from?
Also, when they say researchers pulled the most potent PAH metabolizers, does this mean they have a greater affect on larger hydrocarbons such as something like tetraphene?
Honestly, when I first started reading and found out we were looking at PAH degraders in Antartica it shocked me because I just never assumed anything grew in that region of the world. Knowing that archaea tend to be extremophiles I got to wondering if there was a higher population there over bacteria. I found out that 34% of the prokaryotic biomass was made up of archaea. This leads me to a question about arctic bacteria as well: in the harsh winter season, do some of these adapted bacteria become dormant? The weather can get down to -35 degrees Celsius and salinity of 37-327%.
Building off of this, I think looking at the endogenous and heterologous reporter genes is also very beneficial to the function of the bacteria. The housekeeping genes shows a lot about the metabolism under a variety of conditions. The heterologous reporter genes show the function of the promotor and how that can affect the bacteria.
Looking up upwellings mentions the process of circulating deep water/nutrients to the upper layer. I looked up the process of this in regard to nitrification. It looks like most of the studies are done on warm and oligotrophic open ocean waters. This studied mentioned comparing these upwellings off of the coast of Taiwan to typical coastal upwellings. When looking for the answer to your question, there was a mention about the turbulence of the water. It seems that growth of the microbes is more unfavorable in turbulent conditions.
I agree with this. It is interesting that in this study about upwellings that they are able to survey in the same location, yet the oxygen levels are able to vary. This will lead to less variables in the results than if multiple locations were surveyed.
I believe there is an increased nitrogen and phosphorous concentration in the Black Sea leading to eutrophication. This is due to increased fertilizer use in agriculture and improper wastewater treatment. Cyanobacteria are very prevalent in these waters.
[Total dissolved Mn were determined with inductively coupled plasma and optical emission spectroscopy]
It makes sense that heavy metals are being analyzed as they play an important role in inhibiting the nitrification process. Manganese oxide has a negative effect of nitrifying bacteria. If more of the heavy metal is present, we are most likely going to see a decrease in the amoA gene.
[Sequences retrieved in this study have been deposited in the GenBank under accession numbers EF414229–EF414283.]
I went ahead and went over to GenBank and typed in the accession numbers. I was interesting to look over this under the title of “Uncultured crenarchaeote clone BS100mB1 putative amoA protein-like (amoA) gene, partial sequence” for accession number EF414229.
This is really interesting because it is a natural phenomenon. I also saw that iron can come from the melting of icebergs. It seemed odd to me that iron, a metal, is found in the ocean itself. That is super neat that you talked about this in your geology class, specifically how the dust can travel such a far distance. That seems crucial as majority of the world is made up of the oceans. That reminds me of the Saharan Dust plumes that travels to the coast of my home is Florida. That makes complete sense that the iron comes with that, and I just always thought the sunsets were beautiful!
[the species that produces the siderophore with the highest affinity for iron can enjoy a competitive advantage ]
What happens if the siderophore captures too much iron for the bacteria? Looking at hemochromatosis:the human body absorbs too much of the iron. Is this possible for the bacteria when there is this competition due to the affinity?
I looked this up too. I also was curious what effect recombination had on antibiotics. It turns out that recombination plays a major factor on multi-drug antibiotic resistance. This makes sense as the genetic material is being shared through the genetic material. For chloramphenicol, it turns out the acetylation can prevent it from binding to the target (acetyltransferase).
[15 min for 24 h]
Just a quick comment, but the amount of time that has been involved with this study is crazy. Everything is done so precisely and there are many short times that must be tended to.
[Cell debris was removed from the lysates]
Being new to all of these research techniques, I wasn’t sure why lysing a cell would be beneficial after performing all of the experiments. It makes total sense when you look at it from wanting to maintain the highest yield. How are you able to control specifically what is lysed. After the centrifugation, can further separation be done from the supernatant?
[Geneious]
It’s really neat that there are these resources available now these days that allows for sequencing. This program was founded in 2003. It’s crazy to think about that a lot of these genetic discoveries are not even 100 years old. The discovery of DNA as genetic material was in 1928. The structure of DNA wasn’t even identified until 1953. In my biochemistry class we talk about the protein data bank a lot and it reminded me of that.
Continuing on with the next few sentences: I think the author brings up a good point that with the rich media that there is more than enough iron present within the media. There was exponential growth seen with the minimal marine media, but with the rich media I would predict a constant growth.
I agree with all of this from my analyzation. The increasing concentration of iron results in decreasing siderophore production. This remains consistent with figure 3B with the decreasing relative fluorescence as iron is added. The data also appears accurate as the error bars represent 3 replicates of the standard deviation.
I think it’s so interesting how species are so competitive that they evolve into such a strong species that outdrives the other. Not only is its own self getting stronger through iron uptake, but limits other species to such an extent they can no longer grow.
I was thinking about this too. Where is the limit that causes the bacteria to “switch”. And what if each bacterial cell has a different limit and some are producing while other are cheating.
[the energetic cost to WT V. fischeri ES114 of producing and using these components (Fig. S10A, B). The fact that V. fischeri maintains these functions, despite the cost, further supports the notion that they play a crucial role in survival.]
What are the odds that over time the cell develops a better way to conserve energy. It seems like the common topic in all of my biology courses is to make the most energy possible, but also conserve it. Perhaps an enzyme that could help with this function. I feel like this could lead to an even stronger competitive species.
The deeper the water is, the more less oxygenic. This means that the processes are not as readily available to occur. Therefore, the correlation is that the higher light transmission, the more oxygen is present (closer to the surface).
I think it is the presence of NO2, but also the presence of an oxygen molecules. In a, there is no oxygen present anywhere, but in b and c there is. It makes sense that when NH4 and NO2 are both present 14N15N continues in the upward trend.
Building off of this, it also is interesting to look at the graphs and see the narrow zone of net nitrification as mentioned. It shows that quick and sharp peak seen the strongest in the suboxic zone.
Here to agree with this comment as well. Think about the adaptations organisms have to have in the deep sea environments. Also think about how so much of the ocean is undiscovered. It is hard for us (humans) to explore the deep ocean, which makes sense why is hard for organisms to survive. For sure the nutrient level is a factor. We saw that in some of the previous figures (like why the suboxic zone is the best for net nitrification).
That’s so interesting! I had never heard of any of those techniques prior to this class. Bioremediation is particularly interesting to me. I think this is a field of research that can be incredibly beneficial. Bioremediation could be used to clean up oil spills or possibly target all of the fertilizer runoff into the delta.
I do not see us making much of a difference ecologically if we do not stop relying on oil as our #1 source of fuel for everything. In recent years there has been many efforts to “go green” and be more eco-friendly and while those changes definitely are making a difference, I think ultimately, if we want less contamination and prolonged resources, we need to re-evaluate our current methods. Good news is that research is always being conducted. Maybe someone is our class will figure out a solution for the oil crisis, who knows?
I think it would be really beneficial to collect samples from more than one fuel-contaminated site in order to get the most accurate data representation. In order to get a comprehensive data collection, this research should be conducted annually in order to monitor the abundance of soil bacteria with the growing oil industry. If these samples were collected on the 49th exploration, then studies should continue every exploration there after. The results would allow the researchers to see if the bioremediation that occurs would cause these harmful toxins to decrease over the years. The research conducted in Antarctica can then benefit the rest of the world.
R2A was the selected media for the analysis of this bacteria because of how well bacteria grows and differentiates on it. How would this experiment be hindered if a media was chosen that suppressed growth in some cases? Do you think there is a media that could have been more successful than R2A?
I think we would see less growth on MacConkey (selective of Gram -) and PEA (selective of Gram +). If those agars were chosen, we would be missing out on half of the growth. TSA agar though, I am not quite sure. When I performed a google search, it seems as though most researchers prefer R2A over TSA for a few reasons. While both +/- grow on TSA, there has been roughly a 16% reduction observed in the growth of colonies on TSA compared to R2A. It has also been thought that TSA can be contaminated more easily.
I found this very interesting, here is the link to the paper I read if you are curious: https://academic.oup.com/ndt/article/14/10/2433/1806023
I find this very interesting. It was a bit hard for me to understand with how they worded it though. I couldn’t tell if they used the SSMS approach and it only isolated the most potent PAH metabolizers or if the researchers were just informing us of a new technique that could be used for later study. If this is a technique that was not used, I think this would be so beneficial to re-conduct the experiment using this substrate in order to get a full picture of all of the PAH metabolizers present in the Antarctic soil. While it definitely is great to understand the most potent of the PAH metabolizers, xenobiotics need to be studied more as a whole. With their impact on environmental health as well as human health, more growth in this field is needed to better understand their impact and how they influence the health of the organisms around them.
Hey Rachel,
From what I understand, in situ is different than continuous culturing. When in situ culturing takes place, the membranes allow for the exchange of growth factors and nutrients thus stimulating the bacteria’s natural environment in the culture. In situ conditions would not be steady, they would fluctuate like the bacteria’s natural habitat whereas continuous culturing is a way to maintain constant rates and conditions over an extended period of time.
I was curious as to what a chelator is, so after some research I found that it is a ligand(s) that bind ions/molecules to metal ions. It is often used as a therapy for lead poisoning, but other than that, chelation is widely debated. In this case, the siderophore in V. fischeri binds to the iron present in the water/media preventing V. harvey’s growth. This is a problem because iron is essential in order for V. harvey to grow.
I wanted to see how iron specifically inhibits the growth of these bacteria. Iron deficiency can lead to structual changes in many cells as well as decrease DNA/RNA synthesis. It is also essential is many metabolic processes and products. I found an interesting paper that walks through these processes in more detail if anyone is interested in reading it. https://iubmb.onlinelibrary.wiley.com/doi/pdf/10.1016/0307-4412%2883%2990043-2
The use of both nutrient-limiting and nutrient-rich medias will allow the researchers to elucidate the necessary nutrients that are required in the natural environment in order for the growth and survival of V. fischeri and V. harveyi. This will be beneficial in understanding their development and allow them to monitor their growth under different environmental conditions (i.e. conditions that they flourish in and conditions that they dwindle in).
We use T-tests all the time in my directed studies lab. They are particularly useful to identify significant differences among the data and allows us to compare multiple features to see which pertains to the largest significant difference. Meaning we can compare multiple factors to see which one has the largest influence on the data.
This graph shows the growth of various genes under different settings. We see that in graph A, when 4 of the 5 genes are deleted, production of V. fischeri is greatly reduced, showing that they activate siderophores. To further prove that gene luxT, yebK, fre, and glpK were activators, when placed in the supernatant of V. fischeri is graph B, they grew exponentially because siderophores were present. In graphs C and D, we see that luxT reduces the translational reporter compared to the rest of the genes.
Graph A shows that something present in the V. fischeri culture inhibits the growth of the vibrio species. To begin understanding what exact inhibits their growth, the researchers test the growth of various bacterial species under V. fischeri supernatant. They observe that 3 of the 5 species were inhibited by something present in the V. fischeri. This illustrates the need for further testing.
It is very interesting that an aerE is present in both the cheater and non-cheater. Why do you think aerE would be present if you are not producing siderophores?
I do think you can, however this is definitely a part of the experiment that should be re-done a few times to observe the results. If the growth defect occurs multiple times, I would then say it is sufficient to deem it that possession of AerE makes an aerobactin cheater immune to cytoplasmic aerobactic toxicity.
Yes there is! Oxygen levels decrease with ocean depth, so there is a higher percentage of dissolved oxygen available at the surface. OMZs are found deep in the water column – typically around 700-1000 meters.
I too am very interested to see how this plays out in the rest of the study! I would hypothesize that the OMZs have just enough oxygen available for the aerobic ammonia oxidation to occur and begin the process of nitrification.
I am interested to see if the results reflect that this process does undergo steady state diffusion as the researchers just assumed this. If it is unsteady, that would make for a very interesting scenario for compound fluxes and when nitrification could occur.
Figure 1D shows that nitrification by ammonox bacteria cells occurs within a narrow window of the water column – that window being suboxic conditions. Meaning that the concentrations of Sulfur and Oxygen are present in extremely low levels where nitrification takes place.
This phylogenetic tree displays the species of bacterial and crenarchaeal amoA found at different depths of the ocean. We see that majority of the species were found at 80 meters deep and decreased as we descended through the water column. The * also marks the species uniqueness in comparison to others in that zone. I suppose this is because of the characteristics needed to nitrify at that level under those conditions.
I wonder why we see more diversity within 80 meters as compared to deeper depths. I suppose it is because of nutrient availability. It’s also interesting that BS160B4 was found at 80, 100, and 110 meters!
Figure 1D shows that the anammox bacteria levels are much higher in the suboxic zone. This shows that nitrogen is consumed the most at this level as opposed to deeper depths.
“The degradation of PAH is usually initiated by hydroxylation, especially dioxygenation, which is catalyzed by oxygenase”
Hydroxylation is the 1st step in the process of breaking down PAH’s I was confused before about the difference and or how both process went to start the process. But I did some research outside of the article from https://www.tocris.com/pharmacology/oxygenases-and-oxidases and https://www.sciencedirect.com/topics/neuroscience/hydroxylation and I found that there are several enzymes responsible for the degradation 2 enzymes are a tick for tack situation and are both needed to complete the process.
What did they mean by Island that was kind of over my head did they mean the bacterium was found on an island or it like has a specialized niche in degrading phenathrene?
I understand that both pollutants are in the soil but I wasn’t entirely sure by their wording but are they basically saying that the PAHs are combining with the heavy metals to cause an entirely different substance or are they just saying that both pollutants together are the problem.
I guess another question is assuming that my 1st question was wrong and they aren’t combining with the PAHs even though the Heavy metals are present with the PAHs why are they unaffected by the pollutants of heavy metals?
Last thing I didn’t see them mention a simpler method they didn’t necessarily say what the PAHs needed to maintain their hold on the soil a simple solution to this problem could just be turning the soil exposing the pollutants to the atmosphere or changing the components of the soil which could cause the PAH to degrade through natural causes without microbial interference.
The article stated that “dioxygenase (PAH-RHD) and catechol-2,3-oxygenase (C23O) have been identified as the two key PAHs-degrading enzymes” because they know the problems that could possibly arise from the contamination from the heavy metals can’t they basically do the same thing we are doing with mega and blasting and find where the problem is arise and genetically modify the degrading bacteria to basically side step the contamination assuming of course time, money and that’s the point of interference from the heavy metals.
(in short jus trying to see if a different method can arise to bypass this problem)
2.21 Is essentially what were doing now with our degraders currently. This procedure is I guess easier for me to follow and understand because it’s actually what were doing in lab now. Since its been a little while since 314 this gives me a nice refresher.
I’m just assuming because I’m not entirely sure myself but maybe they were trying to imitate the actual condition of rain and other elements in nature because I wanted to know the importance of the cycles myself.
After taking McCreddie’s biometry class understanding the stats of experiments are easier to understand I wish they would have spent a little more time explaining their work in the Statistical analysis portion of the materials and methods because in reality you can perform any stat to make your data to say what you want it to.
3.2 describes the effects CU(II) had on the degradation capabilities of Sphinobium PHE1 and it goes on to explain that in MSM everything was degraded but it depended on the concentrations the ideal concentration seemed to be less than 600 mg/L.
Rye Grass seems to be the answer in combating heavy metals but as I’ve seen in previous comments plants can take up or absorb metals from the soils, another experiment could be switching plants and trying to determine the variation in plant species.
I see that you have said that you said that we’ve seen 2 of the three paths of the biochemical attack in terms of degradation the mono-oxygenation at the C9 position discusses it’s affects on 9-Fluorenone but is this dealing with the isotope of F I’m not entirely sure.
So just going off the 1st sentence of the final paragraph “dibenzofuran-degrading Terrabacter sp. strain DBF63 can also oxidize fluorene, thanks to a cluster of plasmid-borne catabolic genes.”So are they stating that other PAH degrading bacteria don’t have this gene?
I kind of think that may be the case because even though we haven’t identified all the genes or organisms responsible for PAH degradation but just going off of what I know about bacteria I would assume so because I know there are other organisms from different families would live in the same niche doing the same thing.
Why not use a TOPO Kit? I remember for my MST we attempted a pGEM Easy vector kit but I didn’t receive desired results so I would assume that the TOPO kit is more reliable.
Why not use the 2 step RT-PCR kit isn’t that one considered more accurate, even though it may be possible to introduce more contamination I assume their sterilization and transfer techniques far exceed what we the class would have but they choose the 1 step instead. And I don’t think that contamination was a role player in their choice in terms of 1 or 2 step RT-PCR.
I feel strongly that because of this technique they were easily able to determine family characteristics on how your organism in question would act the phylogeny is basically a way to check and give you a baseline for things to test for.
Fluorine and phenanthrene are the preferred substrates for this microorganism, And based off of their findings even though its not optimum they still can use other PAHs (naphthalene) so can in short I say that all PAH degrading bacteria have the capability of using all PAH’s as a carbon source. I say that because of after the talk a Monday gathered that in the right conditions mutations could allow for strains to pick up the capability of using different carbon sources as energy.
This indicates that this Gram negative bacteria has a entirely new pathway to degrade PAHs. “FlnA1-FlnA2 was shown to be capable of catalyzing monooxygenations and angular and lateral oxygenations of PAHs and heteroaromatics that are not oxidized by DFDO”
So in Short Carbofuran is just a pesticide? I wonder how they decided to observe the effects of this compound and NP degrading capabilities? isn’t this like adding another pollutant to aid in another? That’s how the 1st paragraph seems. The Only thing I could come up with as to why they would even think to use a substance like this to counteract another is carbofuran and it’s organisms don’t last as long as NP.
After continuing reading I understand my misconception with some of the previous statements made in the previous comment. the 1st sentence of the second paragraph set me straight.
Were they indicating that there was a possibility that some fungi have the ability of degrading carbofuran? They identified several strains of Carbofuran degrading bacteria I wonder why they decided to use Novosphingobium KN65.2?
They grew the bacterium on TSB which is the same thing we were using to grow our NP degraders on. Actually a lot of this procedure is very similar to the approach we took to preform our mutagenesis.
So the reason they performed the mutagenesis on tn5 is because that’s where the resistance on km was located, they didn’t clarify that the microorganism was susceptible to km
so summarizing everyone’s comments this strain is basically a specialist that uses carbofuran as a food source with the capability to use other things as food sources
Since PAH bioremediation can clean up pollutants completely or partially, what happens during a partial clean up? Is the area treated with additional clean up techniques to ensure complete removal of pollutants?
In regards to my last question, if the additional elimination technique is used, do they have to wait until the results of the first are shown or can they anticipate the use of a second technique based on the chemical structure of the pollutants in the area?
How was the decision made to utilize only 3 oil contaminated stations? Several samples from several different stations seems like they’d have move data. What systems was used to ensure the marine sediment was successfully not contaminated?
With the research towards the eco friendly solutions, what steps are going to be made? Will they run the process the same way the first was does and what type of degrading bacteria will they add onto next?
The similarity in isolates in this paper compared t0 the random choices of isolates in the last paper seems like the results would be easier to interpret and move forward with. Knowing that certain circumstances were used when choosing colonies would make it easier to decide whether the study went well because of certain phenotypic and genotypic qualities of the bacteria.
Since it is a versatile bacteria, is it possible for it to every work against degrading PAHs and do something different? How interesting would it be if it was able to reverse the work that had already been put in place!
Do these bacteria work best degrading aromatic compounds or does it seem like it is best at almost any compounds since typically aromatics are more complex?
Are the bacteria derived from the hot springs manipulated to perform more closely to archaea? I’m assuming the extreme conditions would favor the growth of a modified bacteria that can withstand the conditions.
Is it the lack of salt in the Bushnell-Haas minimal salt medium that causes the soil samples to become agitated? What significance does this provide to the study??
Were all of the strains identified by similarity to a specific strain? That seems like it would cause errors because the alignments weren’t 100%, at least they weren’t stated to be.
What criteria was used when choosing whether it was similar? In other research based classes there was a certain percentage used as the minimum to choose the similarity.
Would biological methods for treatment of pollutants introduce organisms that could potentially damage the ecosystem? Are the microorganisms selected for specific traits, such as nutrient requirements, from what microorganisms are already present in the affected environment or are they introduced?
Potentially, how would bioremediation occur with a microorganism suited for a highly specific environment? Would genetic alterations be possible to expand the organisms ability to perform set functions or is there a method to synthetically replicate the degradative processes for natural remediation of the pollutants?
What is the ratio of oil stations to area surveyed? Are the islands close with similar environments? Would a more locationally diverse, environmentally similar sampling system give better feedback for the bacteria present?
What is the rate of Napthalene degradation after 15 days? In regard to the experiment, does the degradation continue until all resources are gone without excretion or breakdown that could be harmful to the environment?
What exactly does the bath percentage specify? Also N16, N17, N18 all showed low growth (N16<N18<N17 0.201) with respect other cultures. Yet the emulsification activity varied between N16 and N18 such that N16 was higher than N18. Is it possible that a bacteria could be ill-fitted for growth in the environment but has a higher emulsifying capacity?
Can the conversion of catechols be catalyzed by a pathway other than the tricarboxylic acid cycle? And are the compounds involved in the cycle more or less effective than others?
Bushnell-Haas (BH) minimal salt media is utilized in the examination of fuels for microbial contamination and microbial hydrocarbon deterioration. The media excludes a Carbon-source but contains all other nutrients needed for the selected bacterium.
Within the medium, trace elements include Magnesium sulfate, calcium chloride, and ferric chloride. Ammonium nitrate provides a main nitrogen source. Buffers include mono potassium phosphate and potassium phosphate.
The BH medium can be used at varying pH levels, temperature, and salt concentrations to select for the strains with the highest degradation of hydrocarbon oil.
The liquid Luria-Bertani (LB) medium is a widely used media for the culturing of bacteria. I assume this would be used to grow pure cultures of the bacterial strains with the highest degradation levels in the BH cultures.
Phenanthrene is a polycyclic aromatic hydrocarbon (PAH). It is described as colorless, monoclinic crystal, with a faint odor. Occasionally, they appear yellow. Solutions often exhibit a blue fluorescence. PAHs are formed when oil products (coal, oil, gas, and garbage) are burned but the burning process is not completed.
Bacterial glutathione transferases (GSTs) are enzymes known for their ability to aid in cellular detoxification. They also play roles in protection against chemical and oxidative stress. Due to the chemical composition, they have the ability to catalyze nucleophilic attacks on the electrophilic groups of many hydrophobic, often toxic, compounds.
The C230 sequence is known for its ability in bacterial strains for degrading aromatic pollutants. It is considered an extradiol diogyenase, which incorporates oxygen into one substrate.
The focus on one highly effective PAH degrading strain is unusual in comparison to the last paper where the results of many strains were compared. Is it possible that this study was not as thorough in the broad view of potential PAH degrading strains?
The paper focuses on the relations of the ZX4 strain to S. paucimobilis which is relatively abundant in many environments. Would the study have began with research into the potential heightened PAH degrading qualities of these strains or a selection of strains and then research?
Putative promoter sequences mean the region is believed to function as a promoter but has not yet been proven to be the promoter. What relevance does this have to PAH degraders? Does the proving of a promoter sequence give more insight to how the strain is regulated?
How would the use of the meta-pathway operon aid in improvement of PAHs degradation capability? Could you use gene editing? Otherwise what type of gene modification is being referred to?
A chemoheterotroph is an organism that cannot synthesize intermediates of nutrients and therefore ingests inorganic substances to derive energy from. These types of organisms derive energy from the oxidation of their electron donors, which is why sulfide or H2 are essential.
An anoxic environment is one that has limited or no oxygen available. Therefore anaerobic respiration is most likely to be used to satisfy the microorganisms energy requirements. Mixotrophic organisms should be able to utilize anaerobic respiration and photosynthesis as a means to derive energy. Would phagotrophy or parasitism of any sort be found as well in this type of environment?
As the suspensions bubbled with N2, which is often the end product of nitrogen fixation, can we assume nitrate reductase is active in some or all of these organisms?
The arsenate respiratory reductase genes aids in the conversion of arsenic from a solid form into a usable soluble form. The cycling of arsenic can be influenced by arsenate (As(V)).
Are the lower rates of As(III) oxidation in the presence of light and higher rates of As (V) reduction in the absence of light indicative of the greater energy yielding processes? In light the As(III) oxidation which yields the highest amount of energy most energetically favorable in the given conditions?
Was there a test done in an environment that was strongly oxidizing or weakly reducing? Could that have a significant change persistence of the cyclic nature of the data?
This paragraph discussed the little process of the removal of crude oil from aliphatic components. What exactly is the method of bioremediation? Also, what makes PAHs resistant to biodegradation?
As discussed in this paragraph, the Nakheel beach region did report PAH biodegradation that was not isolated nor investigated. Present times results that there is no evidence of the bacteria strain in that specific region. What could have been the cause of that?
What exactly is the Kimura 2-parameter method? I understand that it is use for a phylogentic tree to establish evolutionary distances. But what is done to contain those results?
PAH degradation I am assuming was used in order to determine the bacterial growth on the NA plates. I understood the other chemicals, but what exactly does inoculum solution do?
Considering that LC grew “poorly” on pyrene and phenanthrene and LB grew on all hydrocarbons except pyrene. Does that make LB is more useful for hydrocarbon bioremediation than LC? Since that it mentions that strains that grew on PAH substrates can be useful for hydrocarbon bioremediation.
Rod bacteria are better than coccoid bacteria because how the rod bacteria contacts with the surface and utilize settled or dense particles. Rod bacteria also divide faster than coccoid bacteria. Is there a way to tell if the population of rod bacteria is greater than or less than the coccoid bacteria?
Interesting to read that the accumulation of HMW can increase the cell weight to where it settles at the bottom of the liquid making it less turbid. What is the purpose of growth assessments?
From looking at the table 1 location A contains less essential nutrients which decreases the productivity. So would location B by looking at table 1 would have the most productivity due to being nutrient rich.
Within the vent fluids are recyclable hydrocarbons? What exactly is the petrochemicals and how is it a suitable natural model system besides that is migrate upward to the sediment surface?
Confuse, so in this studied it talked about studying PAH-degrading bacteria in sediment cores so then how is the hydrocarbons being identified within that?
For colonies forming a clear zones, I assume are the most suitable for the experiment. What exactly does it mean for colonies to form a clear zone, is there PAHs present?
What I got from the introduction paragraph is that selenium when in found in the ocean can be very toxic when there is high levels and can cause severe poisonings to the fish.
The objective for this is experiment is to identify gene that can reduce Se. In past studies Enterobacter cloacae SLD1a-1 is known to reduce Se(VI) and Se(IV).
All the strains were placed on an LB agar, did all of the plates contain the antibiotics? When it says that the “antibiotics were added supplements to the medium” is it meaning extra nutrition for the strains?
The rate of reduction of selenate was calculated by observing its concentration medium. Aliquots of the culture was used in ion chromatography to be analyzed and filtered.
The LB agar is able to identify clones that can perform reduction on Se(VI). White colonies are identified as clones that are unable to reduce, while bright red colonies are identified as clones that are able to reduce Se(VI). In the experiment the results showed that there was no change in color medium with E. coli S17-1, does that mean that the clone did not react, but why was their no white colonies?
FNR is a transcription regulator that is active when oxygen is not present. It makes physiological changes in the cytoplasm. What is the importance of FNR in this experiment
In conclusion of this discussion, that the precipitation of Se(O) can be changed in “”suboxic conditions” by oxygen-sensing proteins. FNR is a potentially control for Selenate reductase genes.
Would this procedure be similar to Sergei Winogradsky enrichment culture technique? Reason I asked this is because they used a selective filter naphthalene to eliminate autotrophic bacteria.
I feel like this is an important part of the research. It could also shield some light on my question I asked in the introduction “What would be the amount of time needed for the bacteria to completely clean the polluted area?” . I think the concentration would have a great effect of the rate it takes to clean the area. Hopefully there will be more about it in the results section.
Based on figure 4 600 ppm or high would be considered harmful to the selected strain because growth at these concentrations are even lower than 2oo ppm which is a fairly low concentration.
Would the gram positive cell wall of the bacteria allow it to be more acceptable in the highly variable environment of oil-contaminated tropical intertidal marine sediment.
As established, 400 ppm is the optimum concentration for napthalene degradation for both gram positive and gram negative. Because gram negative cell wall is more complex would the growth rate of the gram negative bacteria be higher?
Based on the study provided I strongly believe gram negative bacteria would be a great help in cleaning contaminated sites compared to gram positive bacteria.
Because this specific gene is found in different location in different strain, it shows that this characteristic is optional for survival. The reason I believe this it because in one strain the gene is found in the plasmid which carry non essential information and in other strain the gene is found in the chromosome.
Because phnA was not able to convert anthracene and monocyclic aromatic hydrocarbons, that does not have a big effect on its ability to transform other aromatic hydrocarbon such as naphthalene and 1-methylnaphthalene.
When looking at the temperature range and optima for anaerobic As(V) and As(III) other factors should be considered not only light and dark and temperature.
From fig 1 I gathered in light As(III) decrease as As(V) increase in concentration. In a dark environment As(III) increase as As(V) decrease in concentration.
I agree with their detection of oxygen-dependent As(III) oxidation because the sample was collected in a shallow area which is prone to have a high source of oxygen.
Since temperature is a main factor and greatly affect As(III) oxidation and As(V) reduction how does the light and dark contribute to the overall performance of As(III) and As(V)?
Are you suggesting that a colony of bacteria be resistant to different environments all at once? For example, a colony of bacteria could be resistant to an environment with high salinity and high temperatures. If this is possible and the resistant DNA is known for these extreme environment factors, maybe plasmids could be used to splice the DNA into the genomes of the bacteria. However, if the DNA isn’t useful to the bacterial genome, it could later be spliced out.
Maybe multiple simulations were run to test for any potential environmental effects. Also, the products of PAH bioremediation are already present in the environment. They likely won’t cause any new issues.
Is it possible that the different nutrients available at the depths the samples were collected could from could influence genetic mutations in the bacteria collected?
Because the bacteria were cultured in plates with naphthalene as the only carbon source, any bacteria should have been naphthalene-degrading bacteria. Could the bacterial colonies found to have insufficient growth on the initial plate been more successful when cultured on a plate of a different level of naphthalene?
Growth rate and percentage of naphthalene degradation do not appear to be directly correlated at 200 ppm. Strain N7 has a higher growth rate than strain N1, but strain N1 has a higher percentage of naphthalene degradation than strain N7.
It’s most likely that the bacteria they’ve identified can utilize other carbon sources as well. While they can survive using naphthalene as their sole carbon source, it isn’t the only option.
Your hypothesis seems plausible. We had to force our bacteria to degrade naphthalene in lab by reducing or removing other carbon sources. N7 may more readily use naphthalene as a carbon source, especially when naphthalene is the only carbon source available.
It would also be intriguing to see the similarities between the naphthalene degrading bacteria collected in the Persian Gulf and those that could be collected from the Gulf of Mexico.
Based on the text, the claim that they are only partially studied means the bacterium haven’t been extensively researched. Based on the research that was available at the time, they were unable to determine whether marine bacteria or terrestrial bacterial were more efficient naphthalene degraders. I believe this is a way of sort of narrowing the research topic.
DNA is filtered across the agar by size. Smaller pieces travel faster, so the bands visible near the end of the gel farthest from the wells is short sequences of DNA. Bands near the top of the gel are longer sequences.
A low concentrated agarose gel is necessary in this experiment because the DNA sequences are fairly long. This allows for the long but still shortest sequences to travel farther down the gel. A higher concentration of the agarose gel would have likely resulted in wide bands near the top of the gel.
A holoenzyme is expressed if it can breakdown substrates into products. If E. coli is able to do this after the gene has been inserted. The gene should already encode holoenzyme. Genes inserted into a genome in this experiment shouldn’t be mutated.
Transforming the substrates was the test to determine if E. coli had incorporated the holoenzyme into its genome. It also showed how extensively the holoenzymes can breakdown substrates.
Plasmids are nonessential segments of genetic material. While they can often offer the cell antibiotic resistance, bacteria may not necessarily maintain plasmids if the genes are nonessential. Bacteria only need antibiotic resistance when exposed to antibiotics.
The cell likely consumes less energy maintaining multipurpose electrophoresis proteins. Fewer proteins would need to be synthesized, and there would be fewer pathways for mutations.
Based on my understanding, PhnA1 does not code for the small subunit it needs for full monooxygenation activity. Perhaps if combined with PhnA2, it would have a useable small subunit for monooxygenation activity.
There are a number of other amplification techniques that could potentially be useful alternatives to PCR such as multiple displacement amplification, loop mediated nucleic acid sequence based amplification, isothermal amplification, or strand displacement amplification.
What would be the results of an experiment using varying levels of both As (V) and As (III)? The figures depicted only seem to show either As(V) or As(III) as being present and the other being absent.
The breaking of a hydrogen bond leads to higher energy production which is why the Figure 4 shows As(V) being reduced more quickly in the presence of add H2 than in the presence of sulfide. The reduction potential of H2 is more negative.
Water in more shallow areas is constantly being moved. Like in low tide, areas that are usually covered in water are exposed to more oxygen. In deeper waters, the soil or anything growing near the bottom isn’t exposed to much oxygen because the water doesn’t move in the same way.
I interpreted it more as redox rates decreased as toxic byproducts built up, killing some cells. I didn’t think they were ever really measuring bacterial growth rates.
Why is there less information on higher molecular weight PAHs? what does the molecular weight of the PAHS have to do with the degradation? has it never been done or how does it work?
Is the Persian Gulf the only place this can be done? it seems as if it more pollution being done. have they tried anything other than the bioremediation? has it been done effectively to insure more help than harm?
If different zones of the Persian Gulf are polluted why do they keep making it a port for oil. if it had been polluted since the war why keep polluting the gulf? I think more pollution test should be done and I think the bacteria need to be researched due to the fact they may be causing problems as well.
does the ONR7’s various concentrations of naphthalene affect the bacteria growth? does it actually help give results or since it contains naphthalene does it just enhance the bacteria growth of the samples?
why do the aromatic compounds with more rings get decomposed faster? what do the rings actually help in the aromatic compounds if the more you have they are decomposed faster?
how did the gram positive bacteria aid in the degrading of naphthalene in the oil- contaminated topical sediment? does the cell wall allow the bacteria to act accordingly in different environmental settings?
do the harsh/different environments cause the il degrading bacteria? is it created as things take place in different environments? why is it always separated?
since the PAHs are cytotoxic, genotoxic, and, carcinogenic to the marine life, once humans consume the seafood are the PAHs none existent after cooking or are they slowly doing harm to the human body? How can consuming raw marine life or killing marine life that has been affected with the PAHS affect humans?
How do the scientist know when they are placing the specific genus in an area that the area will have the specific carbon sources required to get the degrading done properly?
Indole crystals are very common I see we even use them in lab! but using the crystals do they differentiate the indigo coloring and Naphthalene degraders? and with E.coli not being used as a PAH degrader is this really a smart move?
what is a cosmid clone? why are clones and sub-clones being used in the experiment? will the clones have mostly the same results as the original or does this make the results vary? the lac operon will tell the results of their orient?
If a high concentration of agarose gel would have been used do you think the reults would have been recognizable ? although they would be at the top of the band in a wide sequence do you think it would have been able to be purified?
I also have that question Cheykola! I was wonder if it threw off the results in any way but it seems maybe they drifted from another but still formed together.
Due to the genes having multiple dioxygenase is this good r bad? do we want them to only have one? what harm can be done to the genes or experiment if genes keep being found with multiple alpha subunits?
so do you think if the scientist put down unfit primers in multiple areas, it will hinder the results? do you think they actually start over and put new primers there?
I googled what a NanoDrop spectrophotometer was and I did not know it assessed the purity of DNA as well as quantified it. it allows the samples to be pipetted.
Are clone libraries the same as DNA libraries? DNA libraries are DNA fragments that have been cloned into vectors so that researchers can identify and isolate the fragments.
why do you think the dominant clone group was closest to the 16S? Do you think the reason it possessed the ability to grow anoxygenic photosynthetic oxidation of arsenite from the biofilm but the other clone group was not?
It is weird that the sample could shift whenever the light and dark shifted. how did the sample know when to adjust or when it reverted back the first time why did it keep switching instead f sticking with the last shift it stayed with? is it a special trait the sample possesses that allows it to sense the change?
I looked up what the Cre-lox method was and it is very interesting. It is a site specific recombinase technology. It is used to carry out deletions, insertions, translocations, and inversions in specific DNA sites.
Does it matter or make a huge difference that P73T was grown in artificial seawater? Will this create a difference in sequencing or analysis of the strains?
An HPLC is a High Performance liquid chromatography technique. This is used to separate, quantify, and identify each component of the mixture. It also relies on the pumps to pass a pressurized liquid solvent containing the sample mixture through a column.
This project was much easier to understand. The experimenters seem to know what’s going on and have less variables which makes it less difficult to keep up with.
when I researched this topic it says the name it was under was furadan and it was banned due to the toxicity of this insecticide. it seems no one is using it in cropping fields.
Carbofuran has a very long half life but it is degraded by hydrolysis, if the soil is organic it will speed up hydrolysis which will cause a quicker degradation process.
when it comes to degrading anything the best thing to do is find the best pathways. using liquid chromatography should help because it separates proteins, nucleic acids, or small molecules.
Photosynthetic organisms are photoautotrophs, which mean that they are able to synthesize food directly from carbon dioxide and water using energy from light
The use of the BLAST system to build phylogenetic trees seems to be common. Actually using this technology before allows me to understand why it was used in the experiment and how it helps with protein analysis.
So was the Sphingomonas strain used as a comparison ? As in was it used to compare strains found? Did they use the similarities in other strains to decide they were degraders ?
Horizontal gene transfer is the movement of bacteria from unicellular to multicellular, why do you think the bacteria switched back and forth between the two?
I had to research gene knockout because I had never herd the term. Gene knockout is a genetic technique in which ones of an organism’s genes is made operative.
I know Im slightly late to the discussion, but upon reading through only the introduction and the comments I noticed a lot of people mentioning the BP oil spill in relation to this topic and couldn’t help but think, “Wow we must be responsible for a significant portion of that microbial destruction in Antartica and probably all over the world!” In addition to the oil spills (we know of) the United states is in fact the largest producer and consumer of oil, consuming and producing almost 20 million barrels per day according to the Energy Information Administration of the U.S. (eia.gov). That is an absolute insane amount and the need to shift from fossils fuels and protect the remaining microbial diversity in Antartica and all over the world is an issue that deserves first priority. Just really makes me think how much destruction we have already done that we cannot undo.
I love this commentary Jessica!! In addition to using the proposed solution of using the natural abilities of bacteria, I found it very interesting that although scientists are restricted from introducing foreign organisms to Antartica’s soil under the Antartica treaty, using the native microbes is actually the “best method of choice in the bioremediation of soils with low indigenous PAH-degrading bacteria.” So I also feel it is as you said, “the earth is trying to fix itself/self-heal”and unfortunately we may need to catalyze the process.
This is not something I had not taken into account before when considering the method of PCR. It sounds very logical since DNA has a double stranded helix that is antiparallel so a forward and a reverse primer would be necessary to copy the 16s gene on the corresponding strand. I did a little more research out of personal interest and it turns out The 27F primer would anneal to the antisense of the double-stranded helix (3′ to 5′ direction) and the 1492R primer would appeal to the sense strand of the double-helix (5′ to 3′ direction).
“Polymerase Chain Reaction (PCR).” Diamantina Institute, The University of Queensland, 9 Feb. 2018,
Aliqouts from the four diesel fuel-exposed soils and the four non-exposed control soils were eventually isolated and incubated for 72 hours at 8, 18, and 28°C. Would the Phenanthrene degrading bacteria from the collected sample cultivate more efficiently while incubated in conditions more similar to the environment of King George Island at lower temperatures or would growth be ideal for the bacteria at higher temperatures?
“Bacterial growth in soil from non-contaminated sights were promoted when glucose was added, however not in controlled conditions or when phenanthrene was present. In contrast phenanthrene supplementation of a diesel-exposed soil promotes sudden bacterial growth after 72h, suggesting that phenanthrene is being metabolized and used as a carbon source.”
This excerpt made me wonder how some microbes adapted or were forced to develop in diesel contaminated soils to have the ability to degrade the polycyclic aromatic hydrocarbons . Were these microbes already present in the soil, if so why is growth not promoted by bacteria in the non-contaminated soil using phenanthrene (even if it is a scarcely detectable amount)?
Reading a little further ahead it seems that the polycyclic aromatic hydrcoarbon degrading microbes were present in both samples/soils all along (since 111 were isolated that originated from non-diesel exposed sites out of 350), suggesting that the (PAH) degrading microorganisms flourished over a period of time in the contaminated sites and allowed for exceptional strains of phenanthrene-metabolizing bacteria to develop. The three highest strains from the contaminated soil isolated have the ability to degrade Phenanthrene 69%, 86% and 95% This is a spectacular amount considering the dangerous carcinogenic effect of the (PAH).
I agree with you Jordan. I think the process of microbial bioremediation is the key concept that can be carried over to alternate situations for other endangered ecosystems. The three degrading bacteria genera isolated and observed to be the most efficient degraders of PAHs in this study are restricted to only native bacteria, so there is no telling how many different bacteria with the same ability to degrade PAHs remain undiscovered.
“Several PAH degrading strains of the Sphingomonadaceae family have been isolated from soils from the southern end of the Argentinian Patagonia, ratifying the presence of this type of PAH degrading bacteria in cold environment soils.”
This make me question wether some PAH degrading bacteria are able to live in warmer climates or if any have been discovered. Is the cold environment essential to the process of PAH degradation?
Autumn, I am also unfamiliar with the term “marine oil snow” and thought this excerpt from the University of Delaware did a nice job of explaining this natural phenomenon.
“If you were able to stand on the bottom of the seafloor and look up, you would see flakes of falling organic material and biological debris cascading down the water column like snowflakes in a phenomenon known as marine snow.
Recent disasters like the Deepwater Horizon oil spill in the Gulf of Mexico, however, have added a new element to this natural process: oil.”
Marine snow can include fecal matter, sand, and other inorganic particles in addition to dead animal and plant material. I think that in this case the oil is somehow attaching to these particles, as more accumulates the density increases, and the oil seep down at a faster rate which introduces large quantities of potentially damaging oil to the seafloor.
The deep-sea has received considerably less attention in respect to reports on hydrocarbon biodegradation since the first published reports in 1970. This statement implies to me that there was such an exponential increase in the demand, production, and damage accumulated from oil that we had to dynamically expand research on hydrocarbon-degrading bacteria to keep up with the absolute horrid destruction accumulated from hydrocarbon pollution. Especially after the Deep Water Horizon blowout! Although I think it is beautiful to watch the resolve of the scientific community (such as the proposal/research of bioremediation), it is upsetting that this research stands in our current circumstance as further exploration and production of potentially dangerous hydrocarbons in deeper water continues.
Was 21°C chosen as the second incubation temperature because it is more similar to the average surface water temperature? Or possibly for maximal bacterial growth in the lab?
This was a little difficult to understand for me as well. The internal standard makes sense, but I am unsure about how they would proceed to differentiate between them. u
I suspect it was quite a surprise when the measurements reveled that ”all six of theses PAHs were mineralized by the 4567-24 sediment sample” conversely to the oil rich core 4571-2 which was expected to have yielded higher mineralization levels than that the quite oxidized 4567-24 sediment core.
Ok, so looking at the graph helps to make a little more sense as to how the endpoint selected for extraction of DNA from 13C incubations was 11 days. I can clearly see a progressive removal of PHE occurring around day 9 followed by an asmptote around day 11.
Shelby, this is a great way to set up look at possible future experiments involving these two samples. I would think that the bacterial community from sample 4571-2 would probably be more efficient in mineralization of PAHs in a laboratory environment more similar to its own because of the exposure of oily substances found in the surrounding environment. I think this would definitely be worth pursuing.
I want to make sure I understand this correctly. So essentially of the 68 sequences, 3 OTUs were identified, and OTU-1 comprised the majority of the 68 sequences, so the genus was determined based on the identification of the gene for PHE degradation? Secondly, was this reinforced by the abundance of 16s rRNA genes for SIP clone PHE1 (OTU-1) increasing by magnitudes during incubation?
Wow they already basically remodeled the process of coupled arsenic metabolism under aerobic conditions with an already defined coculture “consisting of aerobic As(III) oxidizer strain (OL1) and an anaerobic As(V) respirer (strain Y5).” I think thats pretty significant and it seems they want to focus this investigation on the phenomena under only anoxic conditions.
It seems Arsenic may be added to this list of biogeochemical cycles of elements as it seems to display the capacity to partake in these coupled reactions that give energy for growth and are essential for some life! I think it is fascinating these microbes can resume a “normal” existence with a metabolic activity that is so alien and it only makes me wonder what else may persist on our planet that could possibly withstand the extremes of other planets.
I was thinking the same thing! “Red-colored biofilms dominated by purple photosynthetic bacteria”? Not only that but it seems that these red biofilms were actually found more common which is quite interesting.
Since the arsenate respiratory genes (arrA) were PCR amplified I am assuming this gene is found in both the archaea and the bacteria since representative clones were sequenced. I was wondering if these genes would be located in similar loci.
Correct me if I am wrong, but in this figure it displays the cooccurrence of Aerobic As (III) oxidation and As(V) reduction both in the presence and absence of light, however the oxidation of As(III) in the presence of light is visibly slower.
As (III) oxidation can be achieved via anoxygenic photosynthesis so I think the fluctuations of these concentrations are a result of the energy source being influenced.
This introduction paragraph explains the topic well and the current research into PAHs. However, it does not clearly state the research that has been done here. The previous paragraph tells us what we need to be looking for in an good introduction, but thus introduction fails to answer the question: What did they do?
I believe that is part of what was explained in class Friday. In order to determine the genes responsible for PAH degradation, we need to be working with a fully sequenced genome. So in this study, where they have isolated samples from various oceans, they must first determine the specimen’s genome before they can determine PAH degradation and genes responsible.
I know that it says that more information is available on all aspects of the mutant generation method, but I wish more detail was given as to how they decided which gene to isolate for deletion. If we do not currently know which genes are responsible for PAH degradation and we are searching for more answers to locating the genes, how did they decide to delete this one in the P73T strain? I would assume more trial and error at several attempts altering genes in other strains before a successful attempt. I would not expect a detailed explanation in this section, but mentioning briefly how it was selected would help to answer, if I plan on conducting a similar experiment, where do I begin to determine which gene to isolate?
Even with your tip, it is difficult to derive too much information from these figures. I understand that the circular maps indicate that there is a GC skew because of the 6th ring in the figure, but are we to deduce any information from the magnification of 60X?
Does the fact that the P73 strain is composed mostly of unsaturated fatty acids also aid in its uptake of aromatic hydrocarbon compound uptake? I would imagine if the membrane is more fluid, then what genes are responsible would be able to control the fluidity of the membrane such that it could allow for uptake of larger hydrophobic substrates as needed.
I wonder if this has to do with the majority of the research involving non aquatic bacteria. If the P73 strain had needed mobility to reach PAHs, it would have had the genetic material to become mobile. However, if the environment were rich enough to produce enough without having to move, it would be biologically wasteful for it to devote so much energy. If the marine environment was indeed rich enough and the current provided all the mobility our bacteria needed for uptake, it could have regressed the mobility adaptation.
Why is it only productive along that route? What happens if 1-hydro-1,1a-dihydroxy-9-fluorenone is not formed? What about the pathway leads to this formation over another?
Thought I would share the results of my wikipedia search for some background for my peers in case they needed some further information on sphingomonads.
The sphingomonads are widely distributed in nature, having been isolated from many different land and water habitats, as well as from plant root systems, clinical specimens, and other sources; this is due to their ability to survive in low concentrations of nutrients, as well as to metabolize a wide variety of carbon sources. Numerous strains have been isolated from environments contaminated with toxic compounds, where they display the ability to use the contaminants as nutrients.
Due to their biodegradative and biosynthetic capabilities, sphingomonads have been used for a wide range of biotechnological applications, from bioremediation of environmental contaminants to production of extracellular polymers such as sphingans (e.g., gellan, welan, and rhamsan) used extensively in the food and other industries.[5] The shorter carbohydrate moiety of GSL compared to that of LPS results in the cell surface being more hydrophobic than that of other Gram-negative bacteria, probably accounting for both Sphingomonas’ sensitivity to hydrophobic antibiotics and its ability to degrade hydrophobic polycyclic aromatic hydrocarbons.
So both gram-positive and gram-negative bacteria are capable of PAH degradation, but fewer data is available on gram-negative bacteria. The reason that there is less information on gram-negative bacteria is because they were until recently believed to be unable to use PAHs as their sole source of carbon?? Is this correct?
Thus, the significance of this study would be to prove gram-negative bacteria capable of oxidizing fluorene and open a greater area of study in PAH degradation.
I was not sure what BamHi, NotI, or NsiI enzymes were so I was reading about them. They are restriction enzymes cable of recognizing short DNA sequences and cleaving them at the target site.
Should be take any special note to the bit about fluorene degradation by protocatechuate in the lower pathway? None of the other information in this paragraph seemed to flow with this bit of information, yet it seemed to stand out to me.
When doing a quick lookup of what protocatechute is the product of protocatechuic acid and O2. Protocatechuic acid is great for pulling off O2 (looking at the structure, it would readily undergo hydrolysis), but the protocatechute did not have much information.
I know that you spoke briefly in class about the insolubility, but I am not sure that I completely followed. I understand heat’s affect on enzymatic activity and that IPTG triggers transcription, but how is it that it goes from being insoluble to soluble?
Does a gram negative bacteria taking on genes from a gram positive bacteria take on more traits from gram-positive bacteria like outer cell membrane? And would that change its stain pick up when trying to stain a sample to determine the cell type?
The table also shows that ability for these pathways to produce alternate products. Do these products need to be taken into consideration when choosing ideal PAH source?
EASYTRAP is a system of recovery of DNA from agarose gel using the principle that glass powder absorbs DNA and allows for simpler and quicker collection of target DNA fragments. It’s more than 50% efficient and uses lower melting temperature.
What does it mean it is realted to another large subunit of PAH dioxygenases but it did not fall in deep branch with other PAH dioxygenases? Either they have left something out here or I am getting hung up on this wording.
The arhA1 gene was inactivated. If the mutant is not involved in PAH degradation, all that is saying is that it doesn’t know the gene’s other cellular functions? I would assume if they knew arhA1 was inactivated, they would no longer be interested in it for PAH purposes.
Is there a way to look at a a comparison between all (or a fraction) of known PAH degraders, their location in the genome. and what PAH they degrade all in one location to be able to visually examine possible connections?
Do all Sphingomonads retain these listed characteristics even if they are the recipient of genetic material from horizontal gene transfer? Or could the obtained genetic material change these “typical” characteristics?
I believe this is the first time we have seen it discussed that there has been an isolation of transcriptional regulators. Would this have been done using RT-PCR to detect expression then genes isolated based on the findings of the expression (in a previous experiment)?
I don’t know that it would be necessary to sequence the entire genome. I think that since the genes involved with the first phase of degradation have already been isolated, they could examine sections of the genome that work in unison with those genes by looking at expression. Creating clones with various sections of DNA which also include the genes involved in the first degradation phase, then plating the clones and examining the colonies for PAH capability, sections of DNA containing the 2nd phase of PAH degradation can be selected for sequencing. This method would be less time consuming and less costly than sequencing an entire genome. Plus I would image it would be difficult to locate the genes encoding the electron-transport of that particular protein even looking at the entire sequence.
Why have they chosen a filter-mating method? I remember reading about this in genetics, but I’m not sure why you would choose this method of conjugation over any other genetic transfer.
Can we break down the logic behind the ligations into the various plasmids? Why did they have to ligate these genes (arhA1A2, arA3, arhA4) into pBBad22T, pBBadA12, pBBadA13, and pBBadA14 plasmids on;ly tp introduce all the plasmids into A4-PCM1? Is it much like the concept of separating the arhA1A2 and ferredoxin/ ferredoxin reductase?
If the P73 gene responsible for PAH degradation was acquired through lateral gene transfer, but the B30 strain did not produce PAH metabolic capabilities, does that imply that P73 acquired that gene through HGT after the 2 strains evolved into different species and now continues to pass the gene onto daughter cells?
Why can they not use a radical reaction or ozone to break the c=c double bond? In organic chemistry, that was the easiest way to break double bonds even with an aromatic. Why can we not throw some O3/Br2/Cl2/ excited electrons in there?
Is it important that they are the only bacteria within one phylogeny family? This is just one family of proteobacteria. Are there other bacteria families capable of similar fluoranthene metabolism?
I honestly would have never known that pollution and oil spills can affect microbial communities. I usually will only think about oil spill and pollution by the beaches and warm oceans. Now that I’m reading just from this paragraph, I can see how oil spills can soak in the soil and destroy small microbiological organisms or destroy fossils. fossils
I am a little confused about the PAH of diesel fuel. Is the different levels of PAH diesel fuel is like the different acidic/ base levels? Is it considered the same chart range we all learned in chemistry? For example, in sentence three phenanthrene is the smallest PAH for the Bay region. This sounds like a PH level of acidic or base level.
Before looking at the comments, I was wondering why does it take so long to analyze the soil too. I was thinking that maybe it takes a while to see bacteria growth from the soil and after it finally grows, they have to separate it to find single colonies. And thinking of that scenario, the scientist have to do that for every base. However, now reading our professor comment, the theory of the length of the expedition makes way more since.
I honestly think it is amazing how these scientist take there time to analyze the culture samples. It surprises me how many steps methods was used to be able to see the colonies in the sample. Hopefully we can do and learn all this in lab just not in a three month time period.
As I am reading towards the end of the paragraph, it says that the scientists found over 350 colonies from the soil count. I just remember from lab 2 where if it was above 250 there is too many to count. I am jus wondering did they count the colonies by hand or was there a computer system that can detect how many colonies? Is tat was it meant by screened, like being ran under a machine?
In this paragraph it says that D32AFA and D43FB had similar growth pattern and used diesel fuel for a energy source. The E43FB did not use diesel fuel for an energy source. My question is what the E43FB use for a source or did it not need a source?
It says in this paragraph that D32AFA and D43FB had the similar growth pattern and diesel fuel was used as a energy source on that site. On E43FB it says that it was unable to use diesel as a energy source. My questions how did the workers use something without diesel fuel or did they use nothing at all?
Now that the scientist have found the three phenanthrene degrading strains, could they find these strains in other cold countries like Canada or cold year round cities like New York? I am asking this due to the sentence that says oil spillage is in cold weather sites. Oil is everywhere?
In this paragraph is it suggesting that scientist who have an interest in this research experiment, do they think that the UBiome company may have used there tested made up samples and not the official samples from the Antarctica sites?
I honestly thought that the oil from the oil spill either disintegrated, got burned off from the hot ocean floor, or cleaned up from the oil rig machines. I didn’t know after all this time oil is still adding to the sea floor.
My question is how do scientist study or collect the hydrocarbon degrading communities without getting burned, the machines getting over heated, or the organisms being burned during that process?
My question is in Figure one, why is it that the sea floor with the oil spill of hydrothermal cores seem rocky or rougher that the regular background core which looks smooth? Does the regular background core have oil spill present also?
I also recognize these primers and using PCR reactions. However, in lab we only did 4 PCR reactions. Its amazing how they are using 96 clones meaning each clone need to be tested with a PCR reaction. That is a lot.
I am wondering why couldn’t authors isolate the organisms from the mid Atlantic Ridge? Was the depth of the sea floor to low to be able to collect a pure organism sample?
As from reading this paragraph, it seems that As(V) and As(III) are dangerous. I wonder what component or chemical nutrient that makes this element poisonous.
I also believe there can be high levels of arsenic in soil all over the world. Especially in countries or states were there are more oil spills or industrial work fields. There can be different levels in a smaller region also. This is what I assume.
I am confused on how does the scientist know the artificial medium will be exactly like the Pahoa Island hot spring water? Do they take a sample from the spring water and compare the chemicals to the medium?
I was wondering since the clones should be a copy of the 16S rRNA gene, how after the blast sequence the clones did not have similarities to the gene database? Is it by the 16S rRNA mutated, and therefore the clone was different than the original gene?
I was thinking the same thing. I was thinking with using the BLAST system in this experiment, every archaea will be slightly the same but, I guess not. Therefore, since there were four different archaea groups did the BLAST tell how did they become different in theory?
I think that is cool too. I wonder even through time and the evolutionary change of the earth will cyclic nature of the phenomenon still be the same or alter in some way?
It is very interesting that they are able to isolate a bacteria from a deep-sea sediment! I am curious as to what made them choose a sample from the Indian Ocean or if this was just random!
This is very interesting to me! I think it is really cool that they are able to grow P73T in the artificial sea water and add in the flouranthene to observe and research it.
As a marine biology major, I am very interested in anything that has to do with the ocean. I have never previously put a lot of thought into bacteria in the deep-sea. I think it would be really cool if we were able to grow bacteria from sediment in the ocean in class! This has made me more interested in the bacterial growth, DNA extraction, and genome sequencing!
I am having a hard time understanding this figure. Maybe because this is the first time I have seen a figure like this. I understand the large one is the chromosome and it explains that from the outside inward is the way the rings are counted. Therefore, is it showing that the very first outer colorful ring and the purple and green ring (1&4) are the forward and reverse non-coding RNA? This figure is just very confusing to me.
This figure shows again the evidence that the chromosome and plasmids have different evolutionary history but in a bar graph form instead of the phylogenetic tree. I like that they used a few different figures in order to show this difference in their evolutionary history.
I have seen several phylogenetic tress throughout my biology classes containing animals/organisms, but this is the first time I have seen one used in comparing gene sequences and proteins. I would have never thought to use this to show data when carrying out an experiment similar to this, but it is a very good way to show this information.
The fact that the P73 bacterium strain could be an aid in any future marine oil spills is very interesting. If this is true, discovering this PAH-degrading bacterium is even more rewarding.
This paragraph states that additional experiments are necessary and more exploration is needed to find more complex information on strain P73. What are these other experiments that will need to be completed?
It is very sad that this contamination is an issue from industrialization and urbanization. The fact that the coexistence of the PAHs and heavy metals can cause damage to the ecosystem and human health should make people want to reduce this industrialization and urbanization. Although most of the human population does not care to do something about this, it is reassuring that it is possible to use the bioaugmentation method to degrade these pollutants without causing other damages and needing too much funds.
I googled what exactly ryegrass is, and it is a very pretty grass. It also said that the ryegrass grows best in mid-winter, therefore, while undergoing this experiment this is a factor they must consider when planting the ryegrass. They must keep it between 5-18 degrees celsius when growing and testing on the ryegrass.
This part of the experiment seems to be the most time consuming taking 84 days just to get the soil they needed to extract the DNA from. Is there any way of speeding this process up? Are the 4 wet-dry cycles over a 4 week time period necessary?
So they created this phylogenetic tree using the same process and program that we used to create our phylogenetic tree in class on January 30th, correct?
These results show that xyLE gene and the C23O gene are more commonly present with a high concentration of PAH, but when the PAH has been degraded those genes disappear. Is this showing that these two genes are involved in the PAH degradation process?
What does the direction of the arrows indicate? I know it says the arrows represent ORFs but it didn’t say anything about the direction of them. I noticed that the arrows for each structure are each going the same direction, but some are just more stretched out or have longer lines between the arrows. This is a figure I haven’t seen before so I wasn’t sure what I was looking at.
In this figure, it shows that the treatment with the PHE-1 and the treatment with the PHE-1+Cu are much lower in PHE concentration than the control. The treatment with just the copper is about the same concentration level as the control which shows the Sphingobium is definitely keeping the PHE concentration down.
In this figure, the expression maintains or becomes level in contrast to figure 2 in which we see the expression of the gene decrease. It is hard to completely compare these figure due to the large difference in the scaling though.
It is very interesting to me that they were able to find out this bacterial strain can help reduce the heavy metal and PAHs co-existing in soils. Although I am curious, is it sure to help with any plant growth like it did with the ryegrass?
Since the genes are dispersed throughout the genome, that makes this species more difficult to study than others. So in the previous two papers we read, the species they studied were not as difficult to determine the PAH degrading genes compared to this one?
The last sentence of this paragraph allows me to understand what they are going to be experimenting in this paper. I was partially confused when reading the first part of this paragraph so I am glad they stated exactly what it is they are going to be looking at.
Thank you, I understand this more now! This is also what we discussed today that we have to find the antibiotic that our bacterium is resistant to in order to complete our random mutagenesis experiment right?
The MEGA software seems to be a very popular software used for alignment and phylogenetic tree construction! This is the software we used and that the past 2 papers we have read use.
Is a truncated transposase a “shortened” transposase enzyme? As in some of the DNA sequences in this gene were deleted? I tried to google this but wanted to make sure I am understanding this term correctly.
Their gel electrophoresis was not a very clear picture. I know they aren’t always perfect, but I think they could’ve got a better picture of their results than the one that is in figure 3.
In the last sentence of this paragraph, they state that the strain LB126 may have been inherited by lateral transfer, this is the same as horizontal gene transfer right?
So for both Sphingomonas and Novosphingobium the catabolic pathway is unclear, but they choose to focus only on the Novosphingobium bacteria in strain in this paper.
This is scary how dangerous the carbofuran pesticide is to humans and other mammals! I googled to see if there were any deaths associated with this pesticide and I read that in Nigeria in 2004 there was one death but several cases of vomiting from the carbofuran residue on batches of noodles.
They state that triplicate cultures were incubated, does this mean they did 3 replicates of each culture? As in 3 replicates of the carbofuran-induced cultures and 3 replicates of the -noninduced?
They grew a culture overnight on agar plates by placing the paper filter onto the agar plates? Then they took the paper filter off the plates and placed into 25 mL 0.01 M MgSO4. I wanted to make sure I am understanding this part of the procedure because I have never performed or seen the filter paper on agar plate techniques.
So the pink to light red color showed up more on medium with just carbon as a source? Or did it still have both nitrogen and carbon but just less nitrogen?
So although this is saying it is closely related to the two different Novosphingobium isolates, that does not mean it has genes that came from those isolates by horizontal gene transfer? Just that it has genes similar to those that are in these isolates?
I can gather from these graphs that as the different mutants are added, the concentration of the formation of carbofuran phenol increases compared to graph a which as no formation. I am not sure what the take home message of these graphs is supposed to be though.
The sentence that states ” The insertions in mutants 6C1 and 21D7 mapped in the same contig.” Is this saying that these mutants did the same thing when inserted?
They decided to test the Novosphingobium to see if it grew on methylamine because the knowledge they had about the first step of the degradation of carbofuran being the release of the methylamine in a different bacteria, correct?
This paper was very difficult for me to understand. So they did find the catabloic genes that aid in the degradation of carbofuran, but they could not identify the genes that are required to initiate the mineralization process?
What physicochemical treatment regimes do biological methods have an edge over? Are in situ biodegradation processes the best way to alleviate pollution even though they can completely destroy organic compounds?
How would results change if the sediment samples were taken from about 50 cm below the surface and seawater samples were not transported to ice but were at RT?
So, one of the main questions with this research is whether gram-negative or whether gram-positive bacteria plays the important role in naphthalene degradation?
Continuing the discussion from paper 1, I know that PAHs are pollutants generated during incomplete combustion of organic materials such as coal and petrol. Bio-remediation is a safe and effective way to eliminate these environmental pollutants by using microorganisms to consume and break down these pollutants.
It would be interesting to see what the phylogenetic trees for this strain looks like and compare to the phylogenetic trees of isolates obtained from the Persian Gulf in paper 1.
In this paper, the Biolog-GN plate was used, but in an article I read, there is another plate that is very similar to this one. The Biolog-ECO plate has a similar use to the GN plate, so why was the GN plate used over the ECO plate for this research? In the article that compared the plates’ ability to distinguish among microbial communities, it didn’t clearly state what the major difference between the plates was. It did state that the two plates demonstrated an equal capacity to discrminate among the micobial communities? Does it have to do with the nutrient media or is there another reason the GN plate was better?
From my understanding, the BH Agar contains all nutrients except the carbon source for bacteria growth. Each isolate was tested in BH liquid medium containing phenanthrene as the sole carbon source, but for the first 7 d, was the phenanthrene the sole carbon source as well?
Nitrification is the conversion of ammonia to nitrate aerobically and denitrification is nitrates being converted to nitrogen gas under anoxic conditions.
What unique characteristic about periphyton made researchers think it would be the right complex mixture for arsenic detection? Was it the components or the function or both?
There is no electron donor in Figure 4 in the top picture. Since the samples that were amended with sulfide decreases, we know that there is sulfur oxidizing bacteria in the biofilm. The samples that were incubated in H2 used arsenate really quickly. The bottom picture shows the consumption of arsenate and regeneration of arsenite.
Can Hydrogen and sulfide serve as electron donors for other growth of diverse arsenate-respiring prokaryotes other than chemoautotrophic growth like chemolithoautotrophic growth that uses C02 as its sole source of carbon for growth?
I do not like one bit the idea of ubiquitous, toxic pollutants tainting the environment which we live. And with potential routes of exposure including via the respiratory pathways, or dermal absorption the issue of these PAHs should no doubt be addressed. I am curious, how are these compounds prevailing so in the environment? Are they being introduced on a huge scale by human activity, or are PAHs forming more rapidly somehow because of an increased level of carbon in our atmosphere?
The genus Rhodococcus sound like bacterial allies in our efforts to remove some of the harmful contaminants maliciously probing about the environment. This key factor of possessing hydrocarbon dioxygenases, detectable in microorganisms by oxidation reactions is fascinating. Such eloquent use of chemistry, and now we have the ability to widely detect and isolate these dioxygenases. I wonder, is there a harmful by-product of PAH degradation?
“However, the genes for the intrinsic electron-transport protein for ArhA1A2, the genes for the degradation pathway of their metabolic products, and their regulatory genes, have not been isolated.”
Should this not be the true purpose of this paper? To isolate these genes?
[By contrast, significant indigo formation was observed only for strain A4-PCM1(pBBadA14) (data not shown). These observations showed that ArhA3 and ArhA4 function as the electron-transport system for ArhA1A2.]
This shows that A4-PCM1 produces indigo from indole, to answer the question you asked.
I personally believe that random mutagenesis is a very important tool that can show us function of a very particular protein structure and it’s function and it could be useful to identify our genes in question.
I think that making mutants is the most straightforward procedure to begin with. Expression cloning and RT-PCR is complicated and requires known primers to conduct the procedure.
[Therefore, strain P73T, the first fluoranthene-degrading Rhodobacteraceae bacterium reported, may be a useful strain in which the C-7,8 dioxygenation pathway involving extradiol cleavage of 7,8-dihydroxyfluoranthene can be studied]
What exactly can this be useful for? Is the dioxygenation pathway particularly important for particular kinds of PAH degradation?
[Notably, region B of the genome, which contained the PAH-degrading genes and were absent in another bacterium of Celeribacter, strain B30, was predicted to have been acquired via lateral gene transfer.]
Maybe I need to look over my genetics and organic chemistry (possibly) but what is the region B in the genome?
To extend upon Shelby’s point about ways bioremediation can be used. Have studies been used to see if these natural microbes can be used in medicine? Has there been any attempts to see how they can decontaminate foreign/mutated cells to a human immune system?
I understand that bacteria and other minerals grow in soil, which is easily found and extracted in warmer climates Though, given hoe such an icy tundra Antartica is, how exactly are researchers able to extract soil samples from ice? Also, what gave them the idea that the natural bacteria microbes from Antartica could be used to decontaminate oil spill in cold water climates?
The phylogenetic tree in this study was constructed utilizing a consensus sequence of the strains. The tree helps to identify certain pathogens that may be of close relation with one another due to ancestral links in DNA. What’s most interesting about this section is the degraders were hydrocarbon instead of PAH as in all of the previous paragraphs.
This is a great question. I too was wondering the possibility of BC1 being utilized in relation to Brucella would have any important correlating consequences for the use in health and agricultural uses.
This paragraph sums up the technique and nutrients utilized to prepare and grow cultures on NA plates. The plates contained diesel, pyrene, anthracene, phenanthrene, or a combination of the 3 PAH as the only carbon substrate. Results showed that certain nutrients and media provided different forms of growth. Isolates LC grew poorly on pyrene and phenanthrene, while LB grew quite well on all media except for pyrene. This test provided information that displayed the ability of the strains to grow a PAH substrates could possibly be useful for hydrocarbon bioremediation.
Find it interesting that rod-shaped bacteria are the dominant morphology, I would expect cocci or spirillum to predominate due to functionality and cross area. This article provides quite meaningful insight into proper function and shape.
The main idea of this paragraph is to inform the readers that certain bacterial inocula have quite specific recipients, and are unable to properly PAH-Biodegrade. It has been studied that bacterial inocula from the same region or similar areas have a higher percentage of effectiveness. Overall this particular study helps to identify, both genetically as well morphologically, the PAH-degrading bacteria from locations such as Eastern Province and Saudi Arabia to determine the limits of their degradation abilities on specific PAHs.
I am still struggling to understand the concept behind this procedure. There seems to be a certain factor behind PAH degraders that does not compute to me. How are the pellets utilized in this experiment?
I agree with Abrianna it is interesting how the difference in temperature can cause a significant difference. Also, what are the components within the cleanup kit?
I believe I see a recurring trend from another article where certain bacteria from the same location display have similar morphologies. In this particular case, they are all gram-negative and rod-shaped. My main concern/ question would be what causes the differentiation in the efficiency of the degraders being that they are retrieved from the same location?
I believe this is an interesting question, the fact that there is a lack of research in this field leaves room for more research to determine the full effects and possibilities for the use of deep water hydrocarbon degradation. Is this possibly a revolutionary discovery or one that can be utilized for decades to come?
How long has the SIP technique been utilized and are there any other techniques available to replace this one that can be more accurate? Also, with depth, do the levels of PAH’s intesify or become less severe.
I was wondering about this as well, I also didn’t see any information related to this topic. As for the microbial mats, I believe this would allow for a quicker cooling time.
I’m not entirely certain of this answer, but I believe it’s to keep the sample from becoming inactive or degraded by the light, which could cause forms of bacterial growth.
I have a similar question, what were the criteria for the selection of the PAH’s and why were there not more than six species chosen for this particular experiment?
What acid was utilized to cause the oxidation to occur in 3 days, also; does this time frame suggest that this occurs at a slow, medium, or relatively fast pace?
Because S17-1 was plated on an LB agar containing tetracycline, which is utilized as a marker for selecting colony vectors. What effect would sodium selenate have?
This may be a simple question but, what does the term cosmid mean when referring to the cloning procedure? Also, how do cosmid clones differ from other types of clones?
This paragraph describes how the wild type fnr mutation was able to restore removed phenotypes. An FNR-like protein was found in E. cloacae which correlated to anaerobic transcription and are an essential protein for the reduction of Se (VI).
If PAHs are formed by two or more aromatic structural configurations, is there a configuration of these structures that is not hazardous to humans. If a person were to come in contact with PAH is there a way to reverse the effects? How long would it take for the symptoms to be reversed?
How many aromatic ring-hydroxylating dioxygenases are there in total? When referring to the multicomponent enzymes, the alpha and beta regions, what are the conserved regions on the alpha particle and how are they used? It’s interesting to learn that aromatic structures play a substantial fundamental role.
It is amazing to me how aromatic hydrocarbons can affect the world in ways such as fossil fuels. How they can be released into the marine environment causing forest or grass fires from petroleum by-products. The fact that PAH’s are cytotoxic, genotoxic, and carcinogenic to marine life and can be transferred to other organisms is interesting to learn.
What causes marine bacteria to be a more significant PAH degrader than the terrestrial habitats? Will there be a larger study on marine bacteria PAH degradation mechanisms? What amount of research was required to determine these facts about obligate marine bacteria, and since there has been limited research, what steps are needed to advance the study and understanding?
OTUs are used to categorize bacteria based on sequence similarity. Each of these cluster is intended to represent a taxonomic unit of a bacteria species or genus depending on the sequence similarity threshold
Essentially, they used the acid to kill the bacteria, so when they incubate it, they are looking for how phenanthrene reacts with other sediments in the sample not just the bacteria. Correct ?
Pyrosequencing is a method of DNA sequencing based on the “sequencing by synthesis” principle, in which the sequencing is performed by detecting the nucleotide incorporated by a DNA polymerase.
It’s really fascinating that PAH-degrading microbes can have enhanced PAH degradation potential by the addition of inorganic and organic supplements. However, will these enhanced PAH-degrading microbes be used in bioremediation? What affect could the inorganic or organic nutrient supplements have on the environment as a consequence?
Toxicity of PHAs are dependent upon a variety of factors such as structure or the way they are exposed in the environment. Which PHA is worse depends on the context of your question. For example, in marine systems LMW PAHs are acutely toxic to marine organisms while HMW PAHs are not. http://ceqg-rcqe.ccme.ca/download/en/243
It is interesting that authors of this study chose to use spread plate as their pure culture technique. Is spread plate more efficient than streak or pour plate under these conditions?
Several times throughout the paper it has been mentioned that cultures are “twice washed and resuspended” in their receptive media. What exactly does this mean? What is the purpose?
Does the YE-supplemented medium stabilize degradation rates that results in a continuous degradation rate with no delay? What caused the sudden rapid phase of degradation on the MSM plate?
Paragraph 12 states that Rhodococcus sp. CMGCZ exhibits potential to degrade PAH Nap, Phe, and Fla as sole carbon and energy sources. Because of this wouldn’t Rhodococcus fit better into set one rather than set two? Despite degrading Fla more efficiently, Rhodococcus still has the potential to degrade Nap and Phe.
It’s really interesting that the YE supplement resulted in the absorption of YE by Nap rather than enhanced degradation. Why did this occur? Is there another supplement known to increase degradation of Nap?
I had a hard time understanding what was meant when the authors described rhodococci as “mycolate-containing nocardioform actinomycetes”. After Googling a bit, I think this means that rhodococci contain salts/esters from mycolic acids ,which are long fatty acids within the cell walls, and that they are branching bacteria that reproduce by fragmentation of filaments.
When would Rodococcus sp. CMGCZ be able to be used for bioremediation operations? Are there certain requirements it has to meet first before being introduced to a polluted site?
With chronic exposure to PAHs the most common effects are symptoms of various types of cancers such as skin, lung and bladder. It’s also interesting to note that its hard to attribute a single symptom to a single PAH because often PAH exposure is by a PAH mixture rather than a single type of PAH.
It’s really interesting that PAHs are often transferred to humans by the consumption of seafood. Don’t let this alarm you, the environmental protection agency has stated that taking in small amount of various PAHs every day will not cause harmful side effects. The FDA also has a method for detecting 15 types of PAHs and their concentration within foods using QuEChERS-based extraction and high-performance liquid chromatography.
For anyone else who didn’t know, sonication is when sound waves are used to disrupt particles in a solution. In this case it is being using to disrupt the cell membrane and release the cells contents. Are there alternative methods for disrupting a cell?
I think you are correct! The aromatic oxygenase gene is what encodes the aromatic oxygenase enzyme which oxidizes subtrates, in this case indole, by transferring oxygen to the substrate.
What affects the activity level of PhnC? What caused the activity of PhnCs with fused, unfused, and monocyclic catechols to the so similar despite having different structures?
The authors make sure to explicitly state that the order of the genes are different from those previously reported for aromatic ring dioxygenases. Can someone help me understand the significance of this?
What causes PhnC to be involved in both upper and lower pathways for degradation of naphthalene, phenanthrene and biphenyl? How does this effect rates of degradation?
From my understanding, FISH technique can be used to identify bacteria in environmental samples. Is it possible that this was the method used or a different one?
This is a great assumption! The PCR we did in lab likely did not go as planned due to the primers not fitting well enough with the genes to produce a PCR product. Maybe it is also possible that the reagents used for PCR were faulty or inappropriate concentrations for the master mix was used. I’m sure this will be elaborated on in the discussion section!
What do the authors mean when they say “all amendments of electron donors and acceptors were made by syringe injection from sterile, anoxic stock solutions?
I did a little research on Euryarchaeota and found out that although this phylum is often found in extreme environments they have also been seen in moderate environments as well!
I think when they author’s state that oxidation and reduction being potentially capable of cooccurence when incubated in light under anoxic conditions means that it is likely that these reactions are coupled as seen in normal redox reactions.
I think we see so few organisms not only because the environment is extreme but because of the size of it. Assuming the ponds they are sampling from are small, there would be a smaller population to reduce competition for resources.
Since we already know that about regularotry genes from other strains of this specie, is it possible that these genes could be similar but just in a different placement in the DNA.
These are the same genes that were done in paper 1 that we read, are they using the mutated strain of A$ to see the degradtion of acenaphthene? Also The regulatory geen clustered tha they isolated, i thougth they already foudn the properties that it was involved in the 1st steps of degrdation of the PAH acenaphthene
WHy were so many plasmids added cotrasnformed? I know they were looking for the particular gene expression, but why woudl they not just put the plasmids in there own ecoli and grow up separatly?
Since they determined where the 4 mutant mini-Tn 5, they used the strain that AG3-69 to look for where they were inserted into the gDNA? or that they were part of the gene encoding for the ferredoxi reductase?
this redA2 homologue is likely involved in the oxygenation of 1,8-NDCA or its metabolites in combination with other oxygenase components, which have not been identified.
is this the gene that they were analyzing in the results section like paragraph 4 i think? that deals with the mutagensis process? Also this gene is important in the oxigenation of the PAH in question, so it should be inhibited if the mutant strain is to not oxygenate the PAH.
In strain A4, arhR is located upstream from arhA3 in the gene cluster; however, the intergenic region between arhR and arhA3 is about 1 kb in length and includes putative transposase genes or their remnants (ORF8 and ORF9). Therefore, autorepression as with typical LTTRs does not seem likely to occur with the expression of arhR.
the LTTRs are not important in the oxygenation of PAHs, or the genes that are expressed to degrade PAH inital step. They are only expressed when the PAH is not present or the genes are not able to be expressed?
CJ2 has had multiple pathways identified in PAH degradation? Has any of the other strain in this paragraph as well? I just am confused on what it was saying a little bit. the genomic island, phn, it tell us about the catabolic pathway that PAH are degraded? is this what other research has been off of too? or did they follow up off the previous papers that we read.
Since this is a new strain, what other bacteria would Strain P73T be closest to when trying to identify the genes for PAHs degradation? Would it coming from the indian ocean maybe have genes that are different from ones already identified?
Why would they choose to do a mutation, if they don’t have the full genome, nor do they have any information of where the oxygenase genes are located on the DNA?
It seems that new genes were discovered for the PAH degradation, so will they have to due a knockout mutagenesis or out in them in a expression vector to make sure they are RHD genes? Does the P73T genes unlock more doors to the wide range of RHD genes that are out in PAH degrading bacteria?
These are 9 genes that are know, but does that mean that if these genes are not present, does that mean that other genes could be there, but the primer did not bond to them?
Why is important that the transporter genes were located in the vicinity of the aromatic catabolic genes? Do they transport these genes? or is this due to like energy source? Also the transmembrane protein genes that were flanking, are these important for PAH degradation?
Why are knowing about these sugars important for the bacteria? Is this for growing up on like a TCA test, which it can use better? I thought glucose would be the best. Also since it is known that they can use these types of pathways, how does this help in the PAH degradation?
[The P73T genome contains 138 candidate genes that may be involved in the metabolism of aromatic compounds, including genes that encode six ring hydroxylating dioxygenases, eight ring cleaving dioxygenases, other catabolic enzymes, transcriptional regulators, and transporters in the degradation pathways.]
Why was further testing not done to help understand these other genes, especially the ones in regionB? I feel like it would have helped with understanding of these newly discovered PAH genomes.
Why did they not isolate the 5 plasmids and do more work with them? I understand it is a genome paper and they discovered some novel genes dealing with PAH degradation, but wouldn’t these plasmids be novel in understanding why these genomes were so important in understanding PAH degradation pathways? I just done understand why they didn’t go any further in their research.
The dissolved selenate was analyzed by ion chromatography. is this method the most efficient for observing this or would an array of methods provide more accurate results?
Isolation by sodium dodecyl sulfate lysis is a form of isolation I am not familiar with. How does this method work and what is the advantage of this method?
The effects of PAHs in the human body is interesting due to their environmental threat. I wonder if the way that PAHs enter our body effect the long term disease that we could experience.
Bioremediation is one of the most interesting uses of microorganisms in my eyes. The positive use of microorganisms to degrade PAHs I’m sure has a huge effect on the diseases that can be caused by PAHs.
I wonder if letting the plates incubate for two weeks instead of 48 hours had any effect on the bacterial colonies exhibiting zone formation. It is interesting that they only incubated CMGCZ for 48 hours after it was picked from the MSM plate, but allowed it to incubate for two weeks to form the zone colonies.
I found it interesting that a delay was observed in Fla degradation for the MSM plates, but not the YMSM plates. I think more information on the chemical properties of the agar plates would help determine this trend.
The difference distinguished between Fla, Phe, and Nap was the opposite of my original thoughts. I thought I would see the exact opposite trend. It is also interesting that the MSM and YMSM plates experience the same difference in degradation.
After reading this entire study and the looking at the molecular weight and chemical composition of Phe, Fla, and Nap, it is still unique to see the degradation potential of Fla compared to the others. Not only would I have hypothesized the results to be flipped before reading this, but I would at least expect the results to be less drastic than what they are.
Looking at the effect yeast has on the degradation potential is interesting to me. Even though the degradation potential of ILCO was not that high, adding yeast as a carbon source still aided in the degrading potential. The results of adding yeast were also experienced in the Fla sample.
With the diversity and capability of rhodococcus, I would be interested to see how its enzymatic effects could benefit humanity. If applied in a pharmaceutical setting, it could make multiple breakthroughs.
I wonder why the substrate was found to not be essential in the degradation rate in the Rhodococcus strain but essential in the R. erythropolis strain. If different substrates were used what would be their effects of the diesel fuel degradation?
If PAHs have the potential to cause cancer, I wonder how often marine life has been at the root of what initiated cancer growth in a human. Are there tests performed that takes away their toxicity before marine life is served to customers?
I’m interested to see the effect of studying a marine bacterium instead of a terrestrial bacterium. I wonder if using these different PAH degraders will have an effect on the overall efficiency.
Determining the function and structure of this set of genes in the Cycloclasticus sp. strain could be critical to determine what genes have significance in the degradation of PAHs. This could allow for the determination of other effective bacterial microorganisms that could be used in the degradation of PAHs.
Interpreting this data and reviewing the methods have helped me to understand the meaning of our work in lab. I now feel more equipped and driven to evaluate my data knowing that our lab work has significance in other experiments.
I wonder if they thought about seeing the effects the chemical purity had on the experiment. I would be interested to see how the purity would effect the overall outcome. I’m assuming it is a significant detail considering the author made a point to mention it.
After doing some research, it was interesting to find out how vital the choice of media is the experiments. We know that ONR7a media is critical for naphthalene degraders because it uses naphthalene as its sole carbon source.
I wonder if the fact that both constructs had a 10.5-kb Sau3AI fragment contributed to the results of both constructs containing an aromatic oxygenate gene. It’s also interesting to me that what we are learning in lab is applicable to these experiments.
I find it interesting that sequencing genomes allows us to see the metabolic properties of these ORFs. I wish the author would’ve told us whether or not this ability to sequence correlates to a specific sequence that can degrade PAHs.
Looking back at the introduction of this paper gave me the thought of if terrestrial bacteria versus marine bacteria had any significance on where the gene clusters were localized. Does the fact that the Pseudomonas sp. strain is a terrestrial bacteria have any influence on its plasmid localization?
What is the difference in the alpha subunit and the beta subunit? What percent similarity was the beta subunit if they both fell outside the major cluster?
I’m interested to see how the outcomes of this study are given. Considering this study will help to further understand the evolution of arsenic from anoxic to oxic conditions.
I’m interested to see why they were not able to obtain PCR products and if it had any correlation to the primer or the enzymes used in the reaction. I’m hoping the materials and methods will reveal what enzymes were use to produce a PCR product.
It is so interesting to me that after reading this entire paper to think about how critical metabolic reactions such as the ones with various arsenic species are in the development of the oxic environment we have today.
Why was it noted that the slurries were stored in the dark? Was the conditions of it being in the dark and held at 5 degrees celsius the reason it was kept viable?
I think it would be interesting to look at the Eurarcheota phylum of Archaea and look at the similarity between the Archaea and the Bacteria found. Maybe they have similar components that could benefit other studies.
It is interesting to see how the oxidation and reduction rates lower each times the light was reimposed on the sample. I wonder if the amount of light used to control this experiment varied at all each time it was reimposed.
It is fascinating to me that in all of the papers we have looked at this semester, the 16S rRNA is foundational part in building a phylogenetic tree. The discovery of the endosymbiotic theory and the conservation of the 16S rRNA has impacted biology tremendously in making these biological connections.
Could the introduction of these multiple bacteria strains have any effect on nutrient levels? or marine life? Like eutrophication for example? or would there not be enough of them to cause an issue?
It is mentioned that the Persian Gulf has a high diversity of naphthalene degrading bacteria. It seems like there is a evolutionary reason for the high diversity due to all the oil traffic interactions in that area.
specifically for the strains that grow on naphthalene. would some of the bacteria we isolated in the previous paper be useful? Since they are naphthalene degrading bacteria and this genus of cyclolasticus can grow on naphthalene.
Does the size of the clusters of ORF’s have any correlation with its degradation ability of PAH’s? I see size related to each other but not efficiency compared with one another.
why does the change in position of the 3-methylcatechol as opposed to the 4-methylcatecol cause such a change in activity? Obviously the addition of a methyl group increases the relative activity for the catechol substrates.
what does a partial ORF consist of? I guess visually I don’t see how this isn’t the 10th and 11th ORF? Does it have something to do with the start codon specifically?
it is interesting that the phnA was able to transform fused cyclic compounds but also unfused ring structures as well. Biphenyl and diphenylmethane are examples of the unfused compounds
It goes much more in depth than simply that but in essence during the day these microbial mats respond differently than they do at night. This has to do with the presence of more oxygen providing more oxic conditions. Where as at night photosynthesis ceases and the mat becomes more anoxic and levels of H2S increase. The organisms respond to the change of conditions and rise or fall.
so I didn’t know what the nylon filters were used for so I looked it up and apparently the nylon filters provide extra stability for the HPLC sample preparation.
I’m interested in what the percentage of acetate was for the 5mM As(v) + 3mM sulfide. Table two says this number was not determined. ( I guess that is what ND is standing for here). I’m Interested because these additions had the highest bicarbonate assimilation numbers. What made the acetate assimilation percentage not able to be determined?
So the halophilic nature of the related Archea would not be present if the clones were take from an area with less salt concentrations? Is that what they are saying?
It interesting to me that something as simple as yeast can help degrade the toxic PAHs. The fact that we find such important usages for such small and seemingly insignificant organisms, like yeast, to help clean our ecosystems is remarkable to me.
What are the differences in the inocula from the different locations? What makes the ones from nearby locations better than ones from far away locations?
Do I need to be able to understand the steps they have described? This section went way over my head. What am I supposed to take away from this section? Because it is bogged down with details I don’t understand.
Doesn’t temperature affect the structure of many key players in biological processes? The main thing that comes to mind is the denaturation of proteins and enzymes at different temperatures. I don’t know if that is what is in play here but it could be along the same lines?
Are they only able to tell the rod shape by looking at the SEM images or is it visible on the agar plates as well? I looked at figure two and the only way I can tell that they are rod shaped is by the SEM images.
So the production of a bio-emulsifier makes the microb a better PAH degrader than just producing a bio-surfactant? Is that for all PAHs or just the high molecular weight PAHs?
In a solution where evaluating the turbidity would show the change in colony population, the solution would be cloudier the colony population increases. Is this only for colonies that are not PAH degraders?
I was confused in the difference between PAHs and naphthalene. I was confused because they seemed to be the same thing but were also used in different parts of the introduction. I googled it and it was cleared up that naphthalene is a type of PAH. It is what is used to make mothballs. It is a good PAH to use in research.
Is the upward migration of the petrochemicals similar to the marine oil snow that occurred after the Deepwater Horizon explosion? Is that what makes this a good model system to study?
Does the warmer temperature of the second sample allow the microbial mats to form or is it the microbial activity and presence that makes the second sample warmer and have hydrothermal influence?
So with all of these genera being PAH degraders, they are found naturally in the deep-sea environment. Are these studies leading to how to increase their PAH degrading capabilities/how to make them more effective? With them occurring naturally and you stating that this means that we don’t have to create superbugs, are we just trying to increase their capabilities?
Is it better for the bacteria to be obligate or non obligate? I am thinking obligate because that means they always degrade the PAHs but I don’t know if I am correct in thinking this way.
So just the location of a gene can change the way an organism functions even though it is the same gene as another organism? I didn’t know the location of a gene could be responsible for so many differences.
So the gene that controls the reduction of selenium is the regulatory gene fnr, but the different forms of reduced selenium are due to the differing enzymes used in the reduction? Does this mean that every selenium reducing microbial has the fnr gene and then different enzymes contribute to the reduction in different organisms?
So three strains were tested and two of them were found to reduce selenium but the genomic library was created for the strain that does not reduce the selenium?
Because we are looking at transconjugants, am I right in saying that this whole experiment is about the selenium reducing gene and not the ability of the bacterial strains to reduce selenium? I want to check because the previous papers all looked at the different bacterial strains doing the degrading.
So to show that they are degrading selenium, they just look for any type of selenium particle that would have been formed from the reduction of selenium? Is there a specific particle they looked for or just any selenium byproduct?
In the LB agar, would the sodium selenate make the agar a differential medium giving the colonies the red or white color depending on if the colony could breakdown the sodium selenate?
so the bacteria will not reduce the selenium until all of the oxygen is used up. is this because it will use oxygen as the electron acceptor until it runs out and then it will start to reduce the selenium by switching to it as the electron acceptor?
So would E. coli be able to reduce selenium if it had the oxygen-sensing transcription factors? is that the only thing holding it back from reducing selenium?
I do agree with your statement about oil being a natural resource that will not be given up any time soon. The paper says, “oil spillage due to transport, storage, and utilization of fossil fuels has become a serious and persistent threat”. So, I think the way we can limit the amount of contamination thereby having no effect on the ecosystem, is if we can find a different way of transporting, storing, and accessing the natural resources that result in less or no spillage. I’m not sure if this is possible since we haven’t figured it out yet. Of course, whatever methods we can resolve must take into account the cold weather and extreme winds of the Antarctica climate.
Hey Abraham, from what I understand after reading an article on Towing Icebergs from Antarctica to the United Arab Emirates in 2018, is that they aren’t sure “exactly how water would be extracted from the iceberg once it reaches the UAE, though the general idea is that the icebergy will be cracked into many pieces, chiseled off bit by bit and loaded into tanker ships which would take the ice to shore and the liquid water that results from the ice melting could then be purified and sold to water companies and government agencies.” Furthermore, 30% of the iceberg would melt during the towing but the amount available once arrived would be between 100 million to 200 million cubic meters of water which is enough to provide around 1 million people with drinkable water for a total of 5 years.
Hey Ryne! From what I understood from the introduction, diesel oil contains PAHs like phenanthrene and it is through bioaugmentation that the researchers “directly seed” those contaminated sites with pollutant-degrading bacteria, i.e. PAH-degraders resulting in what they’re trying to accomplish: bioremediation, which they described as an “economical, and safe approach that can be applied to the decontamination of PAHs with minor alteration of the soil”.
Hey Kamryn! I was also wondering the same thing. I understood that E. coli was used as a negative control as E. coli is unable to grow in either condition. However, it struck me with surprise that there was such a difference in the 2 graphs: with R. erythropolis and S. xenophagum growing more CFU/mL with 0.2% diesel-fuel source vs 0.05% phenanthrene as sole carbon source. Furthermore, P. guineae was extremely drastic in comparison with success in growth using phenanthrene as the sole carbon source but not using diesel fuel.
What would be the significance that the “possibility exists that other strains of interest in our soil samples were not identified”? Are they just saying that since their main goal was to isolate the PAH-degraders, they felt the need to clarify that it wasn’t completely isolated?
[Novel techniques of in situ microculturing have been used to culture Antarctic soil bacteria, by means of a soil substrate membrane system (SSMS), which recreates the native conditions of growth by using the collected soil samples as the nutrient source. ]
Is this similar to a continuous culture or would it be a batch culture or something different altogether? I could be making connections where there are none. I would think it is a type of continuous culture because they didn’t say anything about an accumulation of wastes like in batch culture. Continuous cultures are also better than batch cultures because you can manipulate the amount of nutrients and dissolution rates which would seem beneficial to do.
[The remarkable finding that atmospherically-transported Saharan dust enables proliferation of vibrio bacteria by delivering dissolved iron to surface marine environments]…
This is really cool because in my sedimentology class, which is a branch of geology that deals with the life cycle of sediments and sedimentary rocks, sediments from across the world can travel a really far distance and end up in the oceans through a process called erosion. I never thought about the different heavy metals, like iron, that could be transported along with those sediments.
I noticed in this paragraph (#7) that they state that gram-negative bacteria use a membrane-spanning transporter protein, a siderophore-iron complex, as well as an inner membrane ABC transporter all for the purpose of iron uptake. This was very interesting to me because when we were learning about nutrient uptake and transport in CH. 4, I was under the impression that those processes (ABC-transporters, siderophores, coupled-transport systems, etc.) were separate/independent from each other. I did not know these processes could employ a series of events and co-regulate together.
[E. coli S17–1 λpir was used for cloning, and E. coli MG1655 was used for heterologous gene expression]
I was curious as to what E. coli had to do with experiments involving V. harveyi and V. fisheri, specifically why they used E. coli for cloning and heterologous gene expression, but then I remembered that E. coli is a common model organism in the field of microbiology. I googled the specific reasons as to why E. coli is a model organism and the website cited stated that E. coli has a very simple genome, a fast growth rate, and among other reasons, it is a well-studied organism. Even different strains of E. coli provide substantial contributions which makes sense as to why they are using one strain for cloning and another strain for the heterologous gene expression.
What I understood from this paragraph was that they are using reporter genes to identify whether or not the gene iucABCD is expressed, as this is the function of reporter genes. The fact that it is fluorescent makes it easily detectible in culture. However, will the fluorescence also be easily detectible in the PCR and gel electrophoresis results?
What does it mean that culture fluids containing V. fischeri prevent the growth of vibrio species in minimal marine medium? From what I understand is that V. fischeri is able to uptake the limited nutrients quicker than other vibrio species, consequentially leaving no nutrients left. I also understand that the vibrio growth inhibition only occurs when V. fischeri is added to the culture fluids.
[V. fischeri ES114 culture fluids only inhibited V. harveyi growth when V. fischeri ES114 was cultured in minimal marine medium but not when it was grown in rich medium]
Why is the growth inhibition only present in a minimal marine medium and absent in a rich medium? Is this because the V. fischeri inhibitor molecule works better in a limited nutrient medium whereas, in a rich medium, it is easier for vibrio species to acquire nutrients with or without the inhibitor molecule?
Hmmm, this makes sense because the evolution of siderophore production, i.e. specialized molecules secreted to bind ferric ion and ultimately being reduced into a more useful ferrous form, have contributed to V. fischeri’s acquired competitiveness in its surrounding environment by leaving no iron to the other species.
In paragraph 5 which is what I am thinking Dr. Níhain is referring to when she says “read on. they go through some possible explanations…” they explain that having the AerE gene allows (1.) immunity to cytoplasmic aerobactin toxicity for the cheaters; and (2.) enable aerobactin recycling by the cheaters allowing iron accumulation and acquisition thus increasing their growth.
It seems that the main issue this paper will be focusing on is the genes and enzymes related to aerobic ammonia oxidation, the “first and rate-limiting step of nitrification”. The introduction states that ammonia monooxygenase is one enzyme found in aerobic ammonia-oxidizing bacteria, which is important to the overall research because it is also available in an MGI crenarchaeon, which are likely important and substantial nitrifiers in oceans.
Because of the lack of studies on nitrification along oxygen gradients, this research is being conducted within the Black Sea as the Black Sea is the perfect model. They are trying to identify the marine species responsible for the upkeep of nitrification processes in areas where remineralization, leading to ammonia production and anammox, does not occur.
I think they are analyzing the anammox rates by pairing isotopes because of the fact that isotopes can be used as “bioindicators” and help map out different sources and states of nitrogen (in this case). Since the purpose of this article is to understand better the coupling of nitrification and denitrification/anammox, it makes sense that different isotopes of N are analyzed.
From the PCR they might expect to find evidence of the 16S rRNA in order to determine if the amoA gene is present. This will help them narrow down the nitrifiers within the collection sites of the Black Sea and help them understand the coupling between nitrification and denitrification.
Nitrification levels are basically nonexistent in the anoxic zones and tend to increase in the suboxic zones and then decrease in the same fashion in the lower oxic zones. Furthermore, anammox bacterial cells show a wide variety of activity in the suboxic zones and very little activity in the lower oxic and anoxic zones. As stated in the introduction, substrates from nitrification (a) are used for denitrification processes (d), so it would make sense that A and D have this relation (i.e. as nitrification levels are low, anammox levels are high and vice versa).
(A) shows a high prevalence of crenarchaea cells containing the amoA gene, but a low presence of mRNA copies in the suboxic zone.
(B) shows a low prevalence of the BAOB cells and the mRNA copies in all 3 zones
(C) shows an increasing trend of YAOB cells in the suboxic zones and an alternating increase/decreases trend of mRNA copies between the suboxic and lower oxic zone.
This makes sense because as stated in the introduction, nitrification and anammox have been reported in the lower oxic and suboxic zone, respectively.
[Most of the obtained sequences fell into the marine clusters A, B, and C (20), but three OTUs fell into the “sediment” cluster, which also included Candidatus “N. maritimus.”]
What is the significance of classifying marine clusters vs sediment clusters? I would think the sediment clusters would correspond to deeper depths due to sediment properties–thus, located in 110 m or deeper (i.e. the anoxic zone which is not correlated with nitrification or anammox).
With the help from the legend, when comparing amoA genes found at depths of 80m vs 100m, you can see that there is a majority of species within the phylogenetic tree with amoA genes corresponding to depths of 80m. Furthermore, there is only 1 species with the amoA gene corresponding to a depth of 110. It is interesting that the only species with the amoA gene that has been observed at a depth of 110m has also been observed at 80m and 100m: that species being BS160B4.
I think the purpose of isolating the PAH-degrading bacteria goes back to the introduction where it is talking about microculturing. “New techniques of in situ microculturing have allowed to culture an even broader number of PAH-degrading organisms by recreating native conditions of growth, allowing growth of bacteria not normally isolated using standard culture conditions.” This is essential because they are trying to find the most effective bacteria strains for bioremediation, i.e. the process of safely, economically, and ecologically degrading those harmful PAH compounds.
The purpose of using fluorescence spectroscopy is due to the unique ability of PAH compounds to be fluorescent as stated in the introduction. The use of this type of method of visualizing microbes is better than light microscopy because LM emits a higher resolution allowing you to see the molecular dynamics and interactions in 3D. Thus, if you only used a light microscope, you would not be able to correctly or efficiently quantify the PAH compounds.
Could microorganisms that intake energy from hydrothermal vents help with the biodegradation of oil? If so, this would be an efficient and clean way to help with this problem
Salinity/oxygen levels are differ greatly from the surface of the ocean to the floor. Data samples would be more accurate if they were collected from the floor and surface so that way you could test the differences.
In what way, specifically, does Naphthalene hinder mitochondrial respiration? Are the effects on the mitochondria permanent or once the Naphthalene is no longer in contact with the organism, does mitochondrial respiration resume as normal?
What was the result of the bioremediation in the Persian Gulf? Were the majority of the PAHs bioconverted into micorbial biomass, carbon dioxide, or water or just a fraction? Is there even a need to bioconvert all the PAHs? or will partial conversion be sufficient to make the Persian Gulf “safe”?
Were only the seawater samples transported on ice? or were the sediment samples also transported on ice? Is there a significant reason why the sediment samples were taken at a shallower depth range than the seawater?
What happens to the bacteria when their food source is no longer available? Without a food source would the bacteria just die or would it be possible for them to mutate to obtain food from other sources?
Have there been studies done that prove the resulting biomass is completely benign? What effect does the resulting biomass have on the ecosystem of the effected area?
What was the purpose of filtering the seawater before adding it to the medium? I could see maybe filtering out solid sediments, but if they add the paper back to the medium, which would also transfer the sediments.
If scientists are using bacteria that are already present in the environment how will changing the concentration of those bacteria effect the ecosystem that is present in these regions?
So, it wouldn’t matter if the bacteria had a high
Emulsification activity if it had a low BATH%, since the bacteria wouldn’t be able to adhere for long enough to degrade the naphthalene? Am I reading this correctly?
Would more research need to be done to find strains that can degrade at higher levels? I don’t know the average ppm for oil spills, but I would think that, at least at first, the levels might be higher.
What is the concentration of naphthalene in the Persian Gulf? If it is higher than 400ppm what would the effects of releasing this bacteria on a large scale be?
How does the process of bioremediation, specifically the introduction of large quantities of bacteria, effect the environment? What are the possible side effects of bioremediation in terms of possible decline of other types of bacteria that may be useful to other organisms?
Are all of the PAH degrading bacteria of the same species, or are they different depending on where the pollution occurs? Are all the PAH degrading bacteria close to each other on the photogenic, or have they evolved to be completely different depending on the environment and pollution encountered?
I don’t think they were measuring degradation of phenanthrene, just the bacteria’s ability to use it as the carbon source. I think they were double checking that they hadn’t isolated autotropic bacteria by mistake.
Were these tests done to ensure the bacteria were only able to degrade the phenanthrene? Wouldn’t the bacteria be more useful if they could degrade multiple PAH compounds?
I did a little research, and found out that S. paucimobilis is a pathogenic in humans. How would this affect the viability of using this particular bacteria for bioremediation?
I find it interesting that the bacteria fixed the phenanthrene more quickly at higher concentrations than at lower concentrations. I would think it would be the opposite because lower concentrations would mean there is less to fix which should make the process faster.
What would be the significance of microorganisms being able to use arsenic for energy? Are there areas where arsenic pollution is a problem? What effects do these microorganisms have on humans?
If there are no PAHs available, are these bacteria able to utilize other sources of carbon? I wonder if these bacteria evolved the ability to degrade PAHs as a response to pollution, and if after the PAHs are degraded what the possible consequences of large amounts of potentially pathogenic bacteria in the environment would be.
I would think that they used artificial sea water to ensure that the bacterial strain would grow without possible contamination by other species of bacteria that may be free floating in natural sea water.
I would think selective but enriched. Since it has the sea water in it, and not all bacteria are able to live in environments with a high salt content. It’s definitely a complex medium since we don’t know for sure what exactly is in “yeast extract”
I find it interesting that the strain that they were looking at has the genes for motility but those genes are not expressed in this particular strain. I would think that would be because in water they could just float to where they want to go. It may not be as effective as using a flagella, but it is less energy using for the cell.
I wonder if the strain of bacteria mentioned here doesn’t have PAH degradation genes because the pollution in the Arctic Ocean is not as widespread as in other oceans.
I know there is still pollution in the Arctic Ocean, but I would think it would be minimal compared to, say the Gulf of Mexico. Strains in the Arctic wouldn’t be as exposed to PAHs so the expression of genes for PAH degradation would not be there.
It would be interesting to see what would happen if only the plasmids were knocked out, or if the plasmids were transferred to a different strain of bacteria. The plasmids may not encode metabolic genes but there has to be some sort of use or help that the plasmids give.
I would think having more pathways to use PAHs would be beneficial to the bacteria, since that would allow for degradation of more PAHs. This doesn’t seem to be a problem for this strain though because it says that this strain can degrade many different PAHs.
I agree with Lisa, it’s interesting that they are researching foreign bacteria. I would think that the foreign bacteria would have a larger effect on the ecosystem than using bacteria already present in the soil.
Abiotic stress is the negative impact that non living things have on the environment. This is saying that there are some bacteria that are helpful to plants because the plants are symbiotic hosts to the bacteria. The bacteria get a place to live and things to eat, and are beneficial to the plants by increasing tolerance to pollution in the environment.
I would think it would be advantageous for the organism to have the PAH degradation genes spread through out the genome, rather than all controlled by one operon. That way, the organism would be able to produce different enzymes to catabolize PAHs, rather than just one.
When looking for the genes for PAH degradation, is comparative genomics the most useful part? Otherwise how would they even know where in the genome to start looking?
What happens when the PAH isn’t all the way degraded by the bacteria? What happens to the intermediates when the cell can’t act on them? Are they just expressed as waste by the cell?
Why were the looking at the water soluble-products? I thought that when PAHs were broken down by bacteria for carbon and energy they were completely degraded?
Why was this particular strain picked to do experiments on? It wasn’t one that the researchers had isolated in nature. Was it previously proven to be able to degrade fluorene?
So, in this paragraph they’re saying that the primers originally picked didn’t work in this particular strain. But they picked the primers based on research done on strains that are similar.
Is there a possibility that there are less harmful pathways for carbofuran degradation? I understand that’s not what they’re looking at here, but was just curious.
In this paragraph they are discussing how they came up with the proposed mechanism for carbofuran degradation, right? Were they testing for intermediates in the pathway?
So, if the cfd and cft genes aren’t on the plasmid, then they would be on the chromosome, correct? That would suggest that these genes play an important role other than just metabolizing carbofuran, otherwise it would be energetically expensive to express the genes all the time.
How would researchers involved know that the bioremediation process is an “environmentally benign clean-up?” Are there tests that have proven that the bioremediation process doesn’t potentially cause more harm to local marine life of the Persian Gulf? I googled possible negative effects of bioremediation and had a hard time finding an answer to these questions.
The text states that there were large errors associated with the sulfide-amended samples due to variable kinetics of formation and destruction of various thioarsenic intermediates. Why would these factors have affected the reduction potential of sulfide more than the reduction potential of hydrogen?
I’m confused on why the identity numbers (86/87) for the dominant group (SDT-arrA-13) are significantly smaller than the other Representative clones listed.
Traditionally, lay people view Antarctica as a continent devoid of life. Why would we expect to see bacteria capable of metabolizing diesel fuel and other pollutants in such a desolate landscape? It is easier to understand why bacteria in a place like Nigeria would be more capable of dealing with pollutants, but why would bacteria in Antarctica develop these unique metabolic pathways? I don’t think it would increase their fitness in any way.
I would think that if we are considering microbial bioremediation, the soil would already be beyond natural repair. Would you not expect to have an irreparable change of soil ecology if you are adding a non-native species to tainted soil in the hopes of making it more habitable for native species? I would expect microbial bioremediation to take an extended period of time, and during this time, do we expect horizontal gene transfer to permanently change the gene pool of the native bacteria species?
I understand that bioaugmentation could achieve the same results as introducing a non-native bacterial species to Antarctica in order to deal with pollution. But wouldn’t bioaugmentation bring up the same concerns as introducing a non-native species because bioaugmentation is an inherently unnatural process?
After reading Zachary’s comment, I was curious about the choice of medium. After a colony was successfully grown on M9 minimal media, wouldn’t it be desirable to grow additional colonies from the colony that survived on the M9? Would a more favorable media be chosen for quicker proliferation? Following this line of questioning, is this the next step of the experiment? As I understand it, the goal of this experiment is to isolate diesel fuel degraders to clean up human caused pollution on Antarctica. If a more favorable media was then selected wouldn’t you get more diesel fuel degraders?
Based upon my limited experience in quantitative analysis, multiple extractions are more efficient at removing more of the desired analyte. To answer your second question, I do not believe that the more concentrated analyte should have any measurable effect on increasing the excitation-emission spectra of each sample. I believe the purpose of the excitation emission spectra is to confirm the identity of the excited analyte not how much is there.
The overlap between biochemistry and ecological niches is a very interesting topic to study. Usually when I think of an ecological niche I tend to see it as a place that a species (typically a multicellular animal) occupies and tries to defend from similar competition. It is strange to think of the unseen wars that bacteria wage against each other using the tools of chemistry to outcompete their competition. In many ways it is a much more straightforward mode of competition because of the tools that they use to survive.
I’m curious about the rate of return of the siderophores to their microbial producer. If ferric iron is only scantily present in the culture, as it is in many areas of the ocean, it might be a disadvantage to overproduce siderophores. The wasted energy on the production of siderophores must be putting a severe stress on the vibrio.
Thank you for asking this question. It seems like a great way to get comment points for the most confusing section of the paper. qRT-PCR= quantitative real time PCR. My guess is that used this technique to calculate how much RNA was in the sample. In short they wanted to find out how long the gene was that coded for the selective advantage of V. fischeri.
I’m not 100% confident with this answer but I think that they wanted to stop the growth of the V. fischeri was so they could put it in culture with a polymyxin resistant strain of V. harveyi. In normal conditions V. fischeri outcompetes V. harveyi.
I think it is interesting that Photobacterium angustum was unable to grow in the V. fischeri growth media. It’s strange how a chemical attack by V. fischeri was able to affect multiple different species.
This was the experiment that I worked on in class. I was originally hesitant to post a comment on this section because I did not want to repeat what was stated in the description. My groups take was that figure A shows that the complete deletion of the aerobactin operon would show that no aerobactin would be produced. The more important of the two figures is figure B. Figure B is further proof that aerE is the gene responsible for transporting the siderophore outside of the cell. When aerE is deleting the siderophores are stuck in the intracellular fluid.
In response to your what if statement, I think that each species would have a different limit that tells the organism to either cheat or produce. This would require more research and would be difficult to control because of all of the different interactions present because basically, ecology. I honestly don’t know how an experiment of this type could be set up. Definitely an interesting thought experiment.
The best way that I am able to wrap my head around this section is that vF is trying to monopolize its niche so that vH is unable to displace it. Earlier in the paper it was shown that vH usually outcompetes vF when in a richer media.
The organisms that consume the oxygen are engaging in aerobic respiration and were not the object of the study. I would say that the organisms responsible for the generation of the hydrogen sulfide are the decomposers at the seafloor and also the hydrothermal vents that release a lot of hydrogen sulfide.
I was wondering what natural resources are specifically provided by Antartica. Due to my curiosity, I googled “what natural resources does Antartica have?” I discovered that Antartica has: antimony, chromium, copper, gold, lead, molybdenum, tin, uranium, and zincs. The paper states, “oil spillage due to transport, storage and utilization of fossil fuels has become a serious and persistent threat.” When searching for my previous question, I found out that some people have considered towing icebergs from Antartica in order to provide fresh water to other parts of the world that are in need. If this was to be done, would it do more harm than good?
The paper explains how oil contamination can generate detrimental changes in soil properties. From that, physical and chemical changes promote rearrangements in the soil bacterial communities. This is causing a huge decrease in species richness and soil biodiversity. Oil is a natural resource that we use in everyday life, so I do not think we are going to stop using it anytime soon. With that being said, is there anything we can do to limit the amount of contamination and/or decrease the effect it has on the ecosystem?
The paper states that they used R2A agar when plating the samples. What would the results look like compared to the same samples on TSA, PEA, or McConkey?
What does it mean when it says “cultures were grown with agitation”? Does the excitement of the agitation cause them to grow more rapidly? What is the agitation exactly?
While looking at Figure 4, I noticed that E.coli BW25113 and P. guineae E43FB had low data points in B. However, in A P. guineae E43FB performs higher while E. coli BW25113 is still near the bottom of the chart. What is causing this change to occur?
The paper states that the D43FB strain from Antartica exhibited the highest growth yield. If those same three strains were taken from a different area, such as the Tropics, would D43FB still be the highest yield?
Hey Jacob! I read your comment and thought the same thing, but then I had to know ‘how bacteria do perform in environments like Antartica?’ With that being said I did some googling and discovered that extremophiles known as psychrophiles are known for surviving in cold temperatures. Different species adapt in various ways. However, most trigger a physiological response that allows them to adapt to these unfavorable conditions by making changes in membrane composition. The microorganisms will also adjust translation and transcription machineries. The response from the cold prompts a growth block, as well as repression of translation. Along with this, a set of specific proteins are introduced to make sure the cell metabolism is back in tune and readjust to the new conditions. It also found out that this adaption for E. coli would take about 4 hours.
I know this does not answer your question about the diversity (I am not too sure either) but maybe this is one reason that there can be bacteria diversity in such harsh condition environments like Antartica?
[Microbial bioremediation is quickly becoming an important approach in the continuing efforts to decontaminate critical sites of oil spillage in cold weather environments]
This statement caught my attention because of the “in cold weather environments.” Does this mean that Microbial bioremediation is solely done in cold weather environments, or does it just perform exceedingly better in these conditions compared to somewhere with warmer temperatures? I did some googling and learned that oil pollution in harshly cold weather has led to a higher vulnerability towards the petroleum pollutants compared to tropical and temperate environments. Although I am pleased to acquire this information, my question above still stands. I also apologize if my comment did not clearly reflect my thought process. Basically, I just want to know- is Microbial bioremediation beneficial for other environments?
I am confused with this paragraph. In the beginning it says, “Iron is an essential nutrient for virtually all organisms,” but then states, “iron acquisition presents a challenge to most bacteria.” So, is iron benefitting or hindering bacteria? Does it depend on the situation?
I was interested in the Vibrio species. After I googled it, I found out the siderophores (molecule that binds iron) from these species actually developed through evolution. Since this is a developed system, what did the species do before then to acquire iron?
It never crossed my mind to use iron depletion as a source to induce competition. What other ways are there to promote competition when observing growth in bacteria? How different would this be compared to an environment with an excessive amount of iron?
The scientists screened the colonies for chloramphenicol sensitivity. I did not know what “chloramphenicol” was and had to google it. I found that it is used as an antibiotic for many bacterial infections. It is found in eye ointment and also treats plague and typhoid fever. I think it is neat that chloramphenicol can be used in everyday life products and to help with DNA manipulation and mutant construction!
[For decades, only specific groups of β- and γ-proteobacteria have been found to exhibit this capability. However, recent metagenomic studies in the Sargasso Sea (16, 17) and later of a marine sponge symbiont (18) have identified in marine group I (MGI) Crenarchaeota genes encoding proteins resembling ammonia monooxygenase (AMO), the key enzyme in aerobic ammonia-oxidizing bacteria (AOB).]
Only specific groups were able to exhibit this capability until recent studies. Was this a completely new discovery or did advances in technology help us find this?
I wasn’t sure what “flow cytometry” was, so I googled it. I found that it is a technique used to detect and measure physical and chemical characteristics of a population of cells or particles. It is measuring the volume of cells in a rapidly flowing fluid stream as they pass in front of a viewing aperture.
It is mentioned that gas chromatography was used. This type of chromatography is the process of separating compounds in a mixture by injecting a gas or liquid sample into a mobile phase. It is typically used to test for purity or to separate components of a mixture.
Zachery,
I was looking at the figures and was trying to figure out how to put into words what I thought. The way you expressed this was great on what I was trying to convey!
CARD-FISH is something that is new to me, so I wanted to learn what it was. I found that it is an updated version of the traditional FISH. It is used for phylogenetic staining of microorganisms in various environments. The use of this has helped elucidate the microbial ecology of many habitats.
If Crenarchaeal amoA mRNA variation can explain 74.5% of the nitrite variation within the oxic zone, and AOB amoA mRNA only accounts for 6.5%, what should we expect to be the cause of the remaining 19%?
If bacterial biodegradation is the major contributing factor to cleaning PAH contaminated ecosystems, and PAH’s serve as growth substrates for bacteria; why is bacterial degradation only able to metabolize lower molecular weights? if the bacteria grows in response to PAH’s feeding should it not be able to phagocytize PAH’s of larger molecular weights?
Cyclic aromatic compounds are stated to to afflict great harm to animals and a toxic mutagenic compound which can cause cancer. if these toxins multiply at a very high rate how will bioremediation on its own filter out the toxins? When it is stated in paragraph three line six that bacterial biodegradation is not characterized for high molecular weighted PAH’s ?
The concentration of the turbidity of the water samples obtained to measure the effects of Naphthalene degrading bacteria would be measured by an Assay to analyze the replication of DNA.
gram positive bacteria have a thicker cell wall and can handle the high variations of environmental stress unlike gram negative bacteria. and this can be a factor as to why gram positive bacteria plays an important role in naphthalene degradation.
What were some of the characteristics to colony morphology that you take into consideration upon choosing colonies to isolate. i want to pick up this technique to help me with spotting good colonies in the lab as well.
it was noted int his paragraph that there were physical color changes in in the sample of bacteria, what were the noted color changes and was it considered positive or negative?
How long was the incubation period for the BH liquid cultures, if it was the classical 24-48 hours such as in lab, and there were still no signs of growth then what precautions would you have taken to stimulate the growth of the bacteria. i know in the lab the zinc was added for the nitrate reduction to help the process of negative response.
Open reading frames (ORF) is found in bacteria or archaea genomes. a functional ORF can encode a protein electronically using plasmid pUC119. the process of finding and identifying ORFs begins by the computer finding the start codons and any possible stop codons. the codons are then processed by finding the codon count and possible ribose binding sites. finally the ORFs are organized in a list.
Based on my previous comment the indigo color indicates a gram negative bacteria. from my understanding a Biolog-GN test is a gram negative test is for the purpose of identifying aerobic gram negative bacteria by analyzing the metabolic fingerprint . did you choose the biolog-GN method for fingerprinting the bacteria because it works alongside gram negative results and has a very large number of tests?
Bacteria GST stands for glutathione transferases, in which is an enzyme that participates in detoxification as an aerobic bacteria and is a good method for observing the binding of proteins in an in vitro scenario.
after the oxic and anoxic bacteria has con through incubation under conditions with and without oxygen, afterwards an aerobic and anaerobic oxidizer were each individually added as a defined co culture. oxygen tension meaning different concentrations of oxygenic conditions were used to test its relevance to the bacteria.
the extreme conditions of a hot spring and the gases might closely mimic the environment of early earth an anaerobic conditions, the purpose of this lab might be to find out how the organisms of early earth survived under the extreme conditions
it says that the red colored biofilms were collected through out a large period of time with long gaps in between: August, October, and April. do the climate change in the seasons of these months that the samples were collected not have a variable in the experiment?
i thought the purpose of using N2 gas was to create an atmosphere without oxygen, to resemble the atmosphere of early earth, was the foam stopper the next step to allow gas exchange in small amount to resemble the introduction of oxygen into the atmosphere like that of early earth?
Figure three shows that AsIII is the electron donor and AsV is the electron acceptor. the graphshows a comparison in the temperature ranges Vs. the oxidation of AsIII and the reduction of AsV: AsIII shows a peak at 43 degrees C with a broader pemperature range, and the reduction of AsV shows a narrower temperature range but a comparable optimum.
methanogenisis is a process that is very important to anaerobic breakdown of organic materials in the absence of oxygen to allow the formation of a gas mainly composed of carbon dioxide and methane.
AsIII has a broader temperature range due to the oxidation rate occuring in the dark with no light driven photosynthesis and AsIII is an electron donor in this reaction and a chemolithtroph.
if the springs and the ponds were less shallow and much deeper would the amount of maltonic microbes be fewer and in turn there would be no detection of of aerobic microbes due to the environmental conditions? in this case would the values of AsIII oxidation in figure three be even broader with no peak in temperature optimum?
Why in particular is Antarctica a unique territory in earth’s climate as well as a source of natural resources? It is due to the ecology of the location, climate, natural occurring microbes, etc.
“Oil contamination can generate detrimental changes in soil properties.” Does the rearrangement in the soil bacterial contribute to the growing crisis of climate change which can be seen by the increase in surface temperature, pH, and carbon and nitrogen levels?
To my knowledge, after Googling, 16srRNA has to do with identifying/differentiating between bacteria species. Were certain species of the 350 bacterial colonies selected more prominent than others? Also, did the of acquiring diesel/non-diesel fuel colonies have an impact on which colonies might have an increase in the metabolizing of phenanthrene and were these bacteria related?
Since using a Scanning Electron Microscope is a great way to view biofilms, where their certain colonies out of the diesel/non-diesel fuel which yielded a greater cell adhesion to the phenanthrene? Of these colonies, were there any which made up a greater portion of the biofilm in the “Biofilm Formation Assays”?
“In contrast phenanthrene supplementation of diesel-exposed soil promotes sudden bacterial growth after 72 h, suggesting that phenanthrene is being metabolized and used as a carbon source by the bacteria present in contaminated soils.” What about the chemical structure of diesel-exposed soil allowed the bacteria to utilize the phenanthrene? I tried to google the chemical structure of diesel, and all I found is that it consists of 25% aromatic hydrocarbons, does this play a part in the metabolism of phenanthrene?
“The phenotypic characterization showed positive utilization of substrate for the three stains for D-Glucose, L-arabinose and D-maltose.” Why would the strains not show characterization of L amino acids since that is normally the preferred configuration? Also, if you were to switch the substrates for D-L-D to L-D-L, would the results then show negative utilization of substrate?
It fascinates me that isolate S. xenophagum D43FB not only showed remarkable degradation, but also managed to follow kinetics of growth with other isolates by differing research groups. I would love to know if there are any other regions, beside Antarctica, where this isolate inhabits and would it degrade/replicate at similar rates as it does in Antarctica.
My assumption was the SSMS approach was used to help identify the most potent PAH metabolizers. I do agree with you on “Xenobiotics need to be studied more as a whole.” This was my first time hearing of the term while reading this article. After doing some quick Googling, it astounds me how much impact they have on environmental/human health.
So I googled “direct competition strategies” and found it occurs when individuals are competing for the same resource. If that is the case, are microbes able to utilize transformation of dead microbes DNA in order to gain antibiotic resistance those same microbes exhibit (i.e. production of antibiotics)?
I’m confused by the following “When two or more species each produce siderophores, the species that produces the siderophore with the highest affinity for iron can enjoy a competitive advantage.” Would all siderophore producing microbes not have the same affinity to iron? If not, I guess I’m curious as to what impacts these siderophores ability to “bind” to iron?
“Unless otherwise indicated, erythromycin, chloramphenicol, kanamycin, ampicillin, and polymyxin B were added to final concentrations…” I am confused why the selection of these antibiotics in specific were chosen. Do these play a larger impact when dealing with plasmid introduction?
So I googled Oligonucleotides and found these are short DNA/RNA molecules which have a wide range of applications for testing. I am assuming oligonucleotides are to be used for the “priming” process of PCR. Is there a specific reason oligonucleotides have to be the priming source for PCR?
“We conclude that iron chelation is responsible for the inhibition of V. harveyi growth when V. fischeri ES114 culture fluids are present.” I had to google “iron chelation” and found it is the removal of excess iron through the introduction of special drugs. With this, why would culture fluids from V. fischeri ES114 allow iron chelation to inhibit growth of V. harveyi?
“We conclude that IutA and FhuCDB, encoding, respectively, the siderophore OM receptor and siderophore importer are sufficient to convert a non-aerobactin-producing species into an aerobactin cheater.” This helps answer my question as to “How” non-aerobactin are converted into aerobactin cheaters. However, is there any particular use to being converted to a “cheater” as compared to someone who is already able to utilize iron?
So I had to look up “upwelling regions,” and I found it is when nitrogen-enriched, deep water is moved towards the surface. Is denitrification/anammox more likely to occur at surface level regions as compared to deep level regions due to specific microbes that aid in the nitrification process?
“The first and rate-limiting step of nitrification is aerobic ammonia oxidation.” I am a little confused by this. Since the process is aerobic, it requires oxygen for ammonia oxidation. Yet the Black Sea has low levels of oxygen (I’m assuming the deeper you get into the sea the less oxygen is present) so how are these microbes able to nitrify? Is this why MGI Crenarchaeota is an important nitrifier?
It is interesting to see the abundance of species that are present at 80 m as compared to the other two regions. It appears that as the depth increases, species expressed decreases. I also see BS160B4 is able to be expressed at 80 m, 100 m, and 110 m.
It appears mRNA is able to thrive in suboxic levels in all figures specifically that of Figure C and D. I notice in Figure A, the cells are increasing as mRNA is decreased between 100 m and 110 m.
“It is possible that V. Fischeri ES114 switches between siderophore-producing and siderophore-cheating based on iron availability and whether or not other vibrios are present that can supply siderophores.” The idea of a species being able to “switch” certain characteristics is always fascinating to me. Curiosity just makes me wonder how this species was able to accumulate both producing/cheating traits over time.
“While highly speculative, we suspect that glpK mutation demands that V. Fischeri ES114 use amino acids, rather than glycerol, for growth, altering metabolic pathways that decrease the availability of a substrate required for aerobactin production.” I am a little confused by this statement. What specifically about glpK mutation causes V. Fischeri to utilize amino acids? Also, would this be a beneficial mutation since it “decreases the availability of a substrate required for aerobactin production”?
“Sulfide concentration was measured onboard spectrophotometrically (47).” I remember reading about some bacteria in marine sediments which can transfer the electrons from sulfides and deliver via “chains” to other areas where oxygen is present. I wonder if this is the case here. Maybe the microbes are transferring electrons from oxygen deprived areas to better oxygenated areas of the water? Just a thought.
Brianna, I had the exact same thought. After Googling “Steady State Flux,” it appears to be a constant concentration gradient like you stated. I’m now wondering if this steady state is being used to assist in processes of nitrification.
“Despite the barely detectable gene abundance, strong amoA expression by AOB was detected within the nitrification zone (Fig. 2).” Based on the graph from the previous discussion, this makes sense; however, I wonder if strong amoA expression would be observed in other seas around the world when compared to the Black Sea?
Makayla, I had the same question. I agree it probably has to due with nutrient availability as well as oxygen, pressure, and light available at these depths. I would honestly love to know more about BS160B4. The fact it was found in all three depths was astonishing. I also wonder if it is found else in oceans and exhibit the same characteristics?
If bioremediation is, in part, dependent on the molecular weight of PAHs, would the catabolic activity of biomolecules be hindered or promoted due to a pH change in the environment?
Would it be possible to “condition” naphthalene degrading bacteria in order to allow for habitation of multiple otherwise inhospitable environments? Maybe via selective breeding?
There seems to be a positive correlation between growth and the percentage of naphthalene degradation. This is not a linear trend, because some strains with lower growth have a higher percentage of naphthalene degradation. Could this be due to some strains having a lower volume to area ratio, therefore their growth increases effectiveness?
Because bacteria Cycloclasticus are limited in usability of carbon sources, would they still be the ideal bacteria for aromatic degradation or is it not difficult to keep them alive?
The ability of marine bacteria to degrade more readily could be due to a higher abundance of nutrients for the PAH degrading bacteria to obtain carbon from.
Using both mass spectroscopy and NMR allow identification of the molecule’s molecular mass, which gives insight to stability and reactivity of the molecules. Also NMR gives information regarding the movement of the molecule’s atoms, further allowing for properties of the substance to be interpreted.
Knowing this dominating bacteria are alongside halobacteriacaea indicates the ability to absorb light and use for ATP, but can also function as aerobes.
Why was this bacteria not tested in a usually hospitable water solution that contained arsenic? That could provide more insight to amounts of arsenic/liter this bacteria could reduce.
Their main purpose of this paper was to determine the certain microbe’s use of arsenic in the environment in Mono Lake in order to study HOW these microbes interact with arsenic. The information gathered from this experiment could be used to study and compare the activity in different environments.
Because the Archaea found in this same system are not involved in arsenic cycling, the Ectothiorhodospira are independent when dealing with redox reactions of arsenic and can probably be useful when isolated and placed in a different environment.
Would an environment which supported oxidase genes improve axoB activity? Is there a reason for not testing in an environment which supports oxidase genes?
The differing oxygen levels in marine vs terrestrial environments would play a large role in differences in anaerobic and aerobic pathways in the bacteria, producing different results.
If the strain can grow with fluoranthee as the sole source of carbon and energy, it is able to degrade the compound. What metabolites would researchers be looking for in this situation?
Were these nine genes expected to encode different ring-cleaving dioxygenases based off of previous research or based off of their similarity to one another?
What is the purpose for studying strain B30? Is it strictly for comparison to deep-sea sediment or based off of its likeliness to Celeribacter baekdonensis?
“However, the taxonomic distributions of the five plasmid proteomes of strain P73Twere different from that of the chromosome, suggesting the chromosome and the plasmids may have had potentially different origins.”
This could be due to horizontal (lateral) gene transfer which led to a divergence in its evolutionary history.
Finding out these physiological features of the bacterium would allow researchers to gain a better understanding of how it could be used in real world settings.
Is the identification of the enzymes that govern the initial attack necessary in identifying PAH degrading bacteria, or would it be helpful in the bioremediation using this bacteria later on?
Could the silicone oil be further tested (through Infrared Spectrometry or Nuclear Magnetic Resonance) to determine which PAHs were degraded by the bacterium?
So there is presence of recombinant proteins but they are not active, does that infer the recombinant DNA encoding for the protein was never introduced into the host cell? If so why is this information significant?
Fluoranthene, dibenzofuran, and dibenzo-p-dioxin all yield the highest % product, so these are the best substrates found in the table. Fluorene has a low % yield and produces dihydroxyfluorene, 1-hydro-1, 1a dihydroxy-9-fluorenoned, and fluorene-dihydrodiol. We see that dihydroxyfluorene and 1-hydro-1, 1a dihydroxy-9-fluorenoned are not present in DFDO.
The sphingomonads’ relationship to other pseudomonas is interesting considering it’s more relevant in human diseases rather than wastewater-type treatment of PAHs.
Wouldn’t the inability to oxidize indole to indigo be an unexpected result? And what element of the DNA was unstable? I feel like in the results section that should be classified and then further explained in the discussion.
Strain A4 serving as a substrate assists in the strain’s ability to degrade these PAHs, but was work done to see if they were degraded without the presence of this substrate?
I think finding its homologs would be a great place to start, and then further the research by testing the strain’s ability to degrade these PAHs in an environmental setting.
I believe the purpose of this research was to set up a set of information necessary to carry out further research into the degradation of these PAHs. Other researchers may be able to use these now known PAH-contaminated sites and construct gene probes specific to the sphigomonads in order to leran more about their PAH-dioxygenases.
Do these gene clusters communicate with each other by folding? Otherwise I do not understand how these gene clusters can work together to serve a similar function even with the same sequences.
Glycosphingolipids are involved with signaling between cells. These lipids could be involved with the separate gene clusters working together to serve a function.
ORF 11 being a psuedogene, would there be any further research to determine a possible function of this ORF in a gene that has not been disrupted? Or is it produced solely because of this experiment?
The article cites dermal and respiratory uptake and direct or indirect ingestion as potential ways we can become exposed to PAHs. It goes on to say that prolonged exposure to PAHs can result in cancer of the lungs, heart, or kidneys. This particular fact caused me to broaden my thinking past PAHs to consider what other toxic compounds we can unknowingly come in contact with during our day-to-day lives that put our health at risk.
Jennifer, I too was curious about what caused the change in homology. The ultimate source of evolution is mutation, so I wonder what things in the Rodococci’s environment added up to have an evolutionary effect on the microorganism. With metabolism, growth, and evolution being a universal trend in all organisms, I believe considering the cause of change in Rhodococcus sp. CMGCZ’s homology to be a very appropriate inquiry.
The vast amount of media options available in microbiology are very intriguing to me, but almost quite daunting. I’d be interested to know what steps a microbiologist takes to analyze the different media options in relation to their experiment in order to optimize growth conditions and provide valuable results. Furthermore, does media choice greatly change results, and how do you know if experiment failure/success is a result of media choice or something else?
The paper cites Nap, Phe, and Fla as being the sole carbon and energy sources for this portion of the experiment. What is the purpose of ensuring that the PAHs are the sole carbon and energy source? Is it to ensure that the bacteria are going to seek these out for metabolism and subsequent degradation?
I find it very interesting that the Fluoranthene was completely degraded after one week of incubation. What contributes to my interest in this figure is that none of the others were completely degraded, not even close! I wonder why Fluoranthene was “easier” to degrade than Napthalene and Phenanthrene; I would’ve assumed a low molecular weight PAH, such as Napthalene, would have degraded quicker and more thoroughly than a high molecular weight PAH such as Fluoranthene.
I’m interested in the relationship between time, degradation, and medium choice. More specifically, the fact that the MSM allowed for quicker degradation at the beginning, as opposed to the YMSM which had slow degradation at first followed by a rapid phase of degradation on the 5th, 6th, and final days of incubation. What benefits to bacteria growth/effects on degradation rate does minimal salts medium provide over yeast enriched medium, and vice versa?
I find it interesting that Rhodococcus favors Fla so heavily. Fla is a four membered ring, or in otherwords a High Molecular Weight PAH. With napthalene being a Low Molecular Weight PAH with only 2 rings, I’m surprised that the results supported the conclusion that Rhodococcus sp CMGCZ prefers a HMW PAH, such as Fla., over a lower one, such as nap.. I wonder what contributes to this preference – more carbon, chemical stability of the differing PAHs, etc?
The paragraph states that the Rhodococcus sp. CMGCZ was enriched on Fla in its original growth, and after which was found to degrade 99.3% Fla. It goes on to say that after being continuously subcultured, strains were isolated that were able to grow on Phe. I find it so intriguing how adaptive the bacterium were in order to be able to survive on Phe instead of their usual Fla.!
It seems to me that an organism would prefer to metabolize a more easily degradable carbon source, such as YE, over a difficult one, such as a PAH. If the goal is to use as little energy as possible to carry out the processes necessary to maintain life, then YE seems the way to go over the polycyclic aromatic hydrocarbons. So why then would the researchers in this study choose to provide the bacterium with an “easy way out”?
I’m happy that the paper informed us of how the information gathered from the research would be used. I’m interested to learn more about what specific scenarios and locations these bacteria are used to break down PAHs. Must they be added to an environment, or is the naturally occurring PAH degrading bacteria enough to handle the biodegradation, etc. efforts on their own?
Jennifer, if I remember correctly, the first paper we read discussed a few ways that humans may come in contact with PAHs, one of which being ingestion. This paper cites seafood consumption as a means of ingestion, while the other states that PAHs can be ingested by humans directly or through the food chain. The other ways mentioned in paper one include dermal and respiratory uptake.
Nolan, what an appropriate and necessary thought process! We as consumers need to be made fully aware of what we are ingesting, and what benefits and/or consequences the food in question may bring. I definitely would be interested in learning more about any correlation between PAHs in food and occurrences of cancer. I too would like to know what precautions are taken by food providers.
I am looking forward to reading more about the PAH degradation enabling enzymes possessed by Cycloclasticus sp. strain A5. I hope to be able to compare and contrast the degradation abilities of this marine bacterium and the Rhodococci discussed in the previous paper. I wonder if there is a parallel between the enzymes of the marine bacterium and the ARHDs of Rhodococcus sp. CMGCZ.
I am curious to know the evolutionary history of the genus Cycloclasticus. What environmental conditions and or genetic mutations initially allowed it to degrade aromatic compounds?
Jennifer, I too think it is so cool that we are getting to put into practice what we are reading about in these papers. For this reason, I am very much looking forward to this week’s lab when we get to isolate our bacteria’s DNA! Furthermore, we will get to use the DNA to make phylogenetic connections for our bacterium based on the family of enzymes mentioned in the papers.
I have two questions regarding this portion of the paper:
1. What was the purpose of supplementing with antibiotics?
2. What is the purpose of expressing the gene in a different species of bacteria? Furthermore, why was the lac operon of interest for the study?
I think it is so cool that we are able to determine metabolic properties of microorganisms just by looking at their sequencing. I find it even more interesting that in this study, the phn genes were not clustered as they were in previously reported aromatic ring dioxygenase genes. I wonder how this sets Cycoclasticus sp. strain A5 apart in terms of both evolutionary history and PAH degradation potential. I really look forward to the paper hopefully answering these questions!
I’m intrigued by the phnC gene, as it comes from a subfamily with such diverse sequences. This diversity makes me wonder what happened from an evolutionary perspective to result in such a numerous amount of similar but different genes in different bacterial species. Furthermore, the fact that the enzymes of this group show high activity with different substrates seems promising for bioremediation efforts. Notable aspects of A5’s family, in relation to bioremediation, is their ability to cleave bicyclic substrates.
Madeline, I too was interested by the paper’s mentioning of this difference. It’s strange that A5 differs in such a way from what is “normal” of PAH degrading bacteria. I wonder if the fact that the genes are located on a chromosome instead of a plasmid plays a role in A5’s wide substrate range.
Lane, these evolutionary questions are precisely what I was considering upon reading this section of the paper. The paper cites that the A5 strain has many substrates, and I’m sure this has a lot to do with the overall complexity of the gene cluster we are reading about. Furthermore, I wonder what environmental stressors this bacteria strain endured to have to adapt to multiple substrates in such a way.
It will be interesting to see if and how they experiment with oxic vs. anoxic conditions. After our POGIL in lab today, I am curious as to if this would be a good situation to utilize transcriptomics to look at and compare which genes are active under either condition.
I’m looking forward to reading more about the conclusions drawn from their research. The discussion regarding the role of these metabolic pathways in the Archean Earth is intriguing!
I’m curious about the evolution behind these bacteria’s ability to use arsenic metabolically. Has the species of interest and its ancestors always been able to do this, or was it an evolutionarily advantageous adaptation?
I too thought it was very interesting to think back on our previous papers’ bacteria in comparison to this one. In the previous papers, the research has been utilized for bioremediation purposes. I am curious as to if these bacteria are capable of similar benefits.
I wonder what role sulfide, ammonia, and methane could play for these bacteria. If the arsenite is unavailable which is the most logical next choice? Are these bacteria able to metabolize such properties? I look forward to diving deeper in to metabolism and redox potentials so that I may answer this question myself!
I’ll be interested to see any phylogenetic trees based around these genes presented by the researchers in the results. I’m also interested in seeing if these oxidative genes bear any structural resemblance to those of the previous papers? I know this paper is discussing the metabolism of arsenic and not PAHs, but still.
The paper states that the oxidation rate lowers after each subsequent reimposition of light. Does this occur because metabolic pathways that are more efficient in the dark are being selected for during that time without light, and then out competing others so that when the light is reimposed the bugs present are less efficient at oxidizing AS(III) to As(V)?
In comparing figure 1, anaerobic As(III) oxidation and As(V) reduction, to figure 2, aerobic As(III) oxidation and As(V) reduction, it appears that the reactions are occurring faster under aerobic conditions. I predict this is because the energy generated from aerobic respiration is much higher than that of anaerobic, thereby speeding up the reaction times.
It’s interesting to think about how senescene over time affects metabolic processes across the domains of life. As seen here it dampens the rate of cycling, but this reminded me of how biological aging and mutations affect processes in the human body.
The paper states that different microbial populations play a role in the observable reactions.I think further experimentation and analyzation using metagenomic techniques would be interesting here.
Cyclic aromatic compounds are mentioned as causing significant harm to mammals, however no statistics are given along with this citation. I am interested to know exactly how often or what percentage of the population suffer from exposure to these substances each year. Also, what countries or communites are most impacted by these substances?
Have these microorganisms been isolated in our area and observed in relation to the 2010 Deepwater Horizon oil spill? If not, could this strategy, if effective, also be used to help our own community?
The materials and methods fails to mention how many samples of the bacteria were cultured. Culturing more than one would certainly be important in order to ensure that no contamination occurred and consistent results were obtained.
Is the decrease in naphthalene degradation after 400 ppm a result of sensory overload? If after 400 ppm the bacteria are unable to grow as much then would they eventually be killed by a sufficientally large naphthalene concentration?
I would be interested to see a study that followed the growth rate of these bacteria on a daily basis and in the presence of different concentrations of naphthalene. Would the rate of their growth increase or decrease over time and how would each strains rates differ?
Have more studies been done on this subject at the Persian Gulf since this one was published? If so, are the results consistent and were any gram-positive bacteria ever isolated?
In relation to a previous comment I made, if naphthalene degrading bacteria are present on our own gulf coast in relation to the Deepwater Horizon oil spill, then I wonder if they would be gram-positive or gram-negative. I think that could be a very interesting study to perform with it being so close to home for us.
Bushnell-Haas minimal salt medium is used to determine the ability or inability of an organism to degrade hydrocarbons. This is important for this experiment because it is looking at the degradation of the hydrocarbon phenanthrene.
I looked up and found that sonication is the disrupting of cells using high pitched sound waves. However, I also saw that it is very expensive and usually inefficient. If that is the case, I am curious to know why they chose this method. Was it the only method available for what they wanted to achieve?
Are phthalic acid and diphenylamine unable to support the growth of ZX4 because of their different structures? In other words, the structures are quite different than those of phenanthrene, fluorene, and naphthalene, so is that why they are not useful?
I would think that the lower C230 activity when strain ZX4 is grown only with glucose coupled with the 98.74% of phenanthrene being degraded, indicates that phenanthrene is a better carbon source for it. Is this correct? If so, then perhaps this strain is a good organism to use for bioremediation, assuming it doesn’t have any harmful factors that have not been indicated.
What exactly does the number of base pairs in between the phnH, phnG, and phnI mean? It is also very specific about the number of base pairs upstream from the start codons. What is significant about this fact?
When I looked up Sphingomonas paucimobilis, I learned that it actually used to be a part of the species Pseudomonas. I know these are bacteria that can cause infections in humans. As I search further, I found a paper that looked at the infections caused by this particular strain in patients. Since it is potentially harmful to other organisms, particularly humans, I wonder if the authors of this paper were aware of that when determining it was a good candidate for bioremediation. Perhaps they did, but the strain doesn’t cause enough infections to matter in the long run?
I think I may be getting confused with all the words here, but is this paragraph just saying that the arrangement of these genes is what helped to determine the bacterium the strain was based on the arrangement of the meta-pathway genes?
If I understand correctly, it seems like they are implying that perhaps the genus Sphingomonas may have risen from horizontal gene transfer from the already established Pseudomonas genus. Is this correct?
I looked to see what other toxic elements some bacteria and archaea may be resistant to, and they included bismuth, cadmium, mercury, and several others. I wonder why the authors chose to research arsenic as opposed to one of the other possibilities. Perhaps it was due to ease of experimentation?
When they say “uncontaminated soils” do they mean that the soil is not contaminated with arsenic? If that is the case, then do they offer any theories as to how the organisms developed the ability to reduce or oxidize it?
I don’t really understand what it means when they say they subsampled the slurries each hour with an N2-flushed syringe. Does this mean that a small amount was taken out and reinfected into a new incubation setup?
What purpose does knowing the residual radioactivity serve for this experiment? Is it in order to see the dangers that are still being exposed by the organism even after death?
The way that the organisms are able to switch back and forth between As(III) and As(V) leads me to believe that the pathways and the needs of the organism are being controlled by the presence or absence of light. The figures seem to show that when light is present, the As(III) is being oxidized to As(V), and when light is absent the As(V) is reduced back into As(III).
Could the researchers perhaps take this information and and conducts a more pointed experiment that focuses on just bicarbonate assimilation using more than the two they did here? I just wonder if there may be one that is even better than these two when in the presence of hydrogen and sulfide.
Does this mean that they do not think that Ectothiorhodospira contributed to the dark reaction data at all? If that is the case, then what strains are causing such a high temperature range and rates?
Could we not study the properties of the PAH-metabolism process and form a synthetic version of it for mass use and production? I can see that that would be easier said than done, but it may be a good endpoint to strive toward.
Reading the effects oil can cause on the bacteria living in the soil, remembering the oil spill… Makes me sick. My daughter caught pseudomonas when she was 2 and she will likely have it her whole life, unless of course she receives new FDA approved treatments when she turns of age. Interesting to read that “specially Pseudomonas – dominates over Bacteria that compose the normal inhabitants non-polluted soil.”
I found another similar article discussing King George Island. Doing so I discovered, Different studies on the contaminated soils have revealed the metal is directly interacting with the microbial community. Antartica uses Diesel Fuel as their main source of energy as well as contains 80% of the freshwater reserves of our planet.
Gran-Scheuch, A. (2020, November 7). Effect of Co-contamination by PAHs and Heavy Metals on Bacterial Communities of Diesel Contaminated Soils of South Shetland Islands, Antarctica [Editorial]. Microorganisms.
I had to read more about the KEGG database. I searched and found the database overview which informs readers that it is used as a reference knowledge base for interpretation of large-scale data sets generated by genome sequencing. I also learned, KEGG is being expanded towards integrating human diseases, drugs, and other health related substances. Last updated November 1, 2020
Why not include samples deeper than the surface soil? I would suggest the diesel fuel would absorb into the surface and reach even stronger concentrations deeper into the soil. Transported on ice, makes me wonder if any foreign temperature change would either grow or kill the bacteria.
The soil DNA kit is obviously a good fit for this experiment as they are testing soil from multiple countries, but what other kits would be suited for this lab? Would a different kit have a major impact on the results?
I am unfamiliar with SDS-PAGE GELS so I looked into it. They are used specifically for proteins because the electric currents will only move the proteins based on weight, excluding other factors like charge. The charge of the gel itself blocks the charge of the atoms in the proteins and lowers pH. I also found out that Coomassie blue was an intentional choice as it is formulated work best in this type of gel.
Since the 4-HBA grown cells are more adaptable to different compounds would that make any microorganism with these markers better suited for the degradation of carbon in the areas mentioned in the introduction? Or does adaptability have to do with the influence of other genes?
This sounds very similar to the previous paper based on the research on carbon usage and lack of previous knowledge. I looked into the bacteria being used and it is completely aerobic and has both terrestrial and aquatic species.
I am from Kentucky and it has a major coal/oil mining industry, specifically in the eastern part of the state. Many of the sources of pollution mentioned here are examples of what is being mined across the state. In the region where the most mining is the water is so polluted, there is always a boil water advisory because the treatment plants can’t keep up. I’m curious to know if research like this could be beneficial in keeping contamination levels down.
So far the methods of this paper are much easier to read and understand than the previous paper. Scientific papers, in general, are difficult because of the language used but after doing a lot of these procedures the writing makes more sense. I think I like it more because I know more about the topic.
I was curious about the difference between the gels we have run in class and the ones mentioned here. After some research, I found the gel is similar but the placement in the chamber is different, and therefore the way the electric current flows through it is different. The gel has each end in the buffer but the middle is not covered. The electrodes are on the top and bottom with the buffer and as it runs the lines will move from top to bottom.
Looking at the table I noticed that the % identity of the different orfs are much smaller than what we see in class. Is this due to the type of software we have access to or is it a characteristic of the strain used in this paper?
I am a bit confused about table 2. I see that the last two columns are indicating if each substrate can be transformed by DFDO or CARDO. is the % yield referring to the average when it reacts with both or is it referring to something else?
A lot of the nomenclature in this paragraph is a bit complicated to follow so I did some googling to better see what some of the compounds are. It seems that most are associated with either some type of oils or additives in insecticides or pharmaceuticals. This made more sense seeing as the testing focused on pah degradation.
I also looked into the Glimmer program. It’s used for prokaryotic DNA to find coding and noncoding regions and focuses on long ORFs. It uses a few Markov models to run its data which is different from the other systems.
I agree, I think that the knockout could be useful but a PCR seems to yield a better result with more confidence. I would be concerned about environmental factors or mistakes more in a knockout than in a PCR test.
It seems the B30 strain is very different from the P73 and it does not degrade aromatics as well. It also has less abundant processes and shares no homologs. It’s interesting to see how different species can be even when a part of the same genus.
I knew what HGT was but I was unsure as to how common it is. The paper mentioned how many genes were HTG and I was curious if this was normal or high. I found that in bacteria and other prokaryotes it’s very common for this to happen and will often make up a large portion of the genome.
How rare is it to find the first of something in the gene-sequencing world? I know much of the technology is relatively new so is finding the P73_0346 gene a major landmark or is this common due to the rapidly growing resources?
Could the ability to degrade fluoranthene so well along with the other aromatic compounds be due to it evolving to the environment it was taken from? The Indian Ocean is one of the most heavily polluted areas of the world, especially compared to the B30 strain taken from the Artic.
I want to go into physical oceanography and do research on pollution and or climate change affects. This study focusing on marine based bacteria could very useful in expanding knowledge on how to fight the ongoing issue of mass pollution. If there are natural ways to reduce pollution instead of using man made things with byproducts that are just as bad we can finally find a solution.
I think there’s a lot that can be learned from deep sea sediment and from sediment farther below the surface, especially in regions like this where the water is so old. I think studying bacteria near hydrothermal vents would be interesting as well due to the unique conditions and food sources available
In the previous section I asked if there was a correlation between emulsification activity, cell surface hydrophobicity, and naphthalene degradation and now I see that there is.
I believe it was just worth noting the variation because in the results section it stated that only eighteen showed an adequate amount of growth and were used for more study.
I also did not understand this so I attempted to look up an answer or at least try to gain a better understanding, but I had no luck. Could someone please explain this to me?
In the previous section, I made a comment about sulfide because I did not understand why it was mentioned but now I see that it can be used as an electron donor.
I read that places such as Ireland do routine checks on PAH levels by taking samples of products that are suspected to be contaminated and then analyze them.
I believe the upper pathway is when the aromatic ring is destabilized and the lower pathway results in the production of metabolites that can be used by the bacteria to form biomass.
A monophyletic trait is one that is shared among closely related organisms. And it makes sense that arsenate reduction is not a monophyletic trait. This trait could have possibly been a result of convergent evolution.
This sentence was a little confusing for me. Can someone clarify? I basically understood that isolating the PAH-degrading bacteria that are native to Antarctica can help for treating contaminated soil both in Antarctica and elsewhere in the world. Is that correct?
I’m pretty sure the different levels of PAHs that you’re thinking about are more like different sizes/different number of benzene rings. If you search a picture of phenanthrene, you’ll see it’s a structure of three benzene rings. People are just interested in these because they are a large percentage of what makes up diesel-fuel.
Why was the surfactant production an important thing to test here? Does the production relate to the ability to degrade phenanthrene somehow? Perhaps it affects the ability of the bacteria to swim to its target?
You’re right, it is weirdly like a gram stain. I don’t think it’s meant as a real gram stain because they only used the one dye and never used a decolorizing agent. I assume it just dyed all the bacteria in the culture and they measured the absorbance of the crystal violet to measure the density of the cells in the culture. Almost like a using simple stain.
When it mentions that D32AFA did not differ from the control, it mentions that it means it cannot adhere to the phenanthrene. What are the repercussions of this? Is it possible that this is why it has the lowest degradation of the three strains chosen?
Based on figure 4B, the concentrations of Cd in the Antarctic soil would render the microbes practically useless for degrading phenanthrene. I’m surprised that wasn’t mentioned outright in the results. I wonder if it’ll be in the discussion.
I calculated the lowest degradation % in Figure 4B to be 5mg/Kg if were assuming 1mL= 1g. Considering the concentrations in the snow was 15-85mg/Kg, it’s unlikely that the microbes can degrade at all in the Antarctic soil.
I think that the extreme environments require the microbes to be more innovative with their uptake of nutrients so researchers are more likely to find the unique metabolic properties in those areas.
In order to decontaminate the oil spillage areas through bioremediation, do they just plan on growing specific microbes in a lab and dropping a lot in the area to clean it up? Similar to probiotics?
So in this case of SIP, would they be creating oil with an isotope that is different from everything else they give to the microbes and see if the isotope ends up in the cells?
[…mineralization assays using 14C-labeled…]
What’s the difference between performing mineralization assays using a heavier isotope and SIP? I thought that’s what SIP was.
I understand the need for an internal standard, but if you’re adding unlabeled E. coli DNA to a mix of enriched and unenriched sample DNA, wouldn’t the E. coli DNA get confused with the unlabeled sample DNA and possibly mess up the measurements? With measurements as small as 5 microliters, it wouldn’t be very accurate.
Or was this separation not measured and used specifically for visualization?
When I originally looked at Table 2, I was confused to see 4567-24 showing higher mineralization yields than sample 4571-2. I’m glad they touched on why that may have occurred. Would it be realistic to see a future study where samples were taken in the same location as 4571-2 and tested using the same method except under anaerobic conditions?
So, on the DGGE, the top bands are the ‘heavy’ DNA and the bottom are the ‘light’ DNA, and the fact that samples 6-10 show just ‘heavy’ where 12-17 show primarily ‘light’ DNA shows that the DNA was properly separated through isopycnic ultracentrifugation. Then they used samples 6-10 for further analysis because they had the ‘heavy’ DNA.
Is that correct?
I agree. They took the 4571-2 microbes from a hot, pressurized, sulfide-rich environment, then tested them under conditions better suited for the 4567-24 microbes. I’d like to see another experiment with conditions more similar to the 4571-2 environment.
Agreed. It seems so much harder to get energy out of PAH’s than other carbon sources. They must not have anything else around to take from if they lost their ability to get energy from other sources.
Does the soil in Japan naturally have higher levels of arsenic in it? Is that why even uncontaminated soils have microbes that are capable of this? Or is this normal, and they were just giving an example from Japan?
I believe so. They figure out the chemical makeup of the water, then recreate it under lab settings to ensure it stays the same and stays sterile throughout the experiment. I’ve done it before with tapwater experiments.
Either that or the abundance of microbes would adjust so the microbes that make As(III) would be more abundant than the ones that use it in order to maintain the concentrations.
I’m curious why the temperature range was wider at night than during the day. I expected it to be lower because it’s colder at night. I guess maybe it’s just circumstantial that it also deals with high temperatures better? Or maybe I just don’t know enough about hot springs to make an educated guess.
It’s cool to see that they can use either hydrogen or sulfide as electron donors for their reactions. It’s even cooler that we now understand why hydrogen works so much better for microbes as an electron donor!
This paragraph sounds like there needs to be a lot of future research on differentiating the bacteria in these samples. They can tell you that some bacteria are similar to previously published ones, but they can’t tell you which ones or even confirm that it’s the bacteria of interest that are related.
PAHs are then transformed through different peripheral pathways into a few key intermediates (such as protocatechuate, salicylate, gentisate, and catechol)
Have we talked about these before? I don’t think I remember these in the previous papers, either.
I think I am confused as to why it seems that they are studying this bacterium and this specific PAH when they seem to have already pointed out how similar they are to their respective counterparts.
Regulatory genes are often transcribed in the opposite direction. They are often on separate transcriptional units (makes sense given that they control the transcription of other genes) and having them transcribed in the opposite direction helps ensure that they are regulated independently of the genes they regulate.
They want to examine the roles of particular combinations o genes so they have to subclone them. Look at the gene map figure to see which pieces they’re putting together.
” However, it was recently reported that the LysR-type transcriptional regulators (LTTRs) ThnR, PcpR and LinR, which activate the expression of target degradative genes, were isolated from Sphingomonas strains that degraded tetralin, pentachlorophenol and γ-hexachlorocyclohexane, respectively.”
I read the other section connecting to this and cannot understand the purpose of including this sentence. Can it be explained?
Ashlyn commented: This portion basically answers my first question, but I am interesting in seeing exactly how many genes are involved in the degradation process.
“However, the genes for the intrinsic electron-transport protein for ArhA1A2, the genes for the degradation pathway of their metabolic products, and their regulatory genes, have not been isolated.”
In the sentence after this, they talked about using transposon mutagenesis for isolation except for that of Arh1A2. Why is this?
“We also investigated the regulatory mechanism of a newly isolated gene cluster, which includes the arhA genes, as the first step for elucidating the entire acenaphthene degradation system in strain A4.”
I thought in this particular kind of bacteria the genes were scattered throughout the genome. Were the genes clustered together in the lab, or am I getting this confused with something else?
I’m not sure why acenaphthene is less studied than naphthalene, phenanthrene, and pyrene. It’s on EPA’s priority pollutant list. It may be that it’s more difficult to obtain isolates and that the “true” PAHs are easier to study and a better model for aromatic hydrocarbon biodegradation. If you look up the MSDS for acenaphthene and compare it to naphthalene, they’re pretty equivalent.
Most bacterial groups other than the sphingomonads co-locate their PAH degradation genes. The sphingomonads are unusual in that their PAh biodegradation genes are not all located together in the chromosome. The sphingomonads tend to have a lot of small groups of PAH genes but don’t usually have the entire pathway at the same location. It makes finding the genes for the entire pathway more challenging than for say, pseudomonads. One explanation for this is that the sphingomonads may undergo more genomic rearrangements than other genera of bacteria.
That’s not really important. Focus on why they did the experiment. What did they hope to achieve by doing transposon mutagenesis? Were they doing random or targeted mutagenesis? What genes were they hoping to disrupt and why?
They did two separate disruption experiments, one where they knocked out ORF 15 and a second where they knocked out arhR yielding two mutant strains A4DR (no arhR) and A4D15 (no ORF 15). THe direction of transcription is irrelevant for the knockout as they did the knockouts separately.
Why do you think they chose to knock out these particular two genes?
Here they are expressing the dioxygenase genes (arhA1&A2) with and without the A4 electron transfer proteins (ArhA3 and ArhA4) to determine which gene products are necessary for dioxygenase activity. Remember that in our first paper they used electron transfer proteins from another Sphingomonas strain and did not get any indigo formation when they expressed arhA1A2 alone.
By amplifying across 2 genes they get to see if the genes are transcribed as one polycistronic message (controlled by one promoter) or if they are transcribed individually (each from a separate promoter). This experiment allows them to assess regulation of the acenaphthene degradation genes.
See the first paragraph of this section – Gm is short for gentamicin, an antibiotic. The miniTn5 transposon they used for random mutageneisis contains a gentamicin resistance gene as a selectable marker. All mutated A4 colonies will acquire Gm-resistance when the miniTn integrates into the A4 genome.
Kristen is correct. They did mutagenesis and some mutants lost the ability to convert indole to indigo. This ability is a proxy for the first step in PAH degradation. The mutants that can no longer do this must have a mutation affecting the initial step in the pathway.
Mutants that no longer cleared acenaphthene from the plate could have a mutation anywhere in the pathway before the metabolites enter central metabolism.
You need to think through/remember what you learned about PAH metabolism in the previous papers.
They know which genes are missing in the mutant and which ones are still present. By adding back some of the missing genes they can assess their function more accurately than in E. coli as all the other acenaphthene degradation genes are present and should be transcribed and translated correctly by A4’s genetic machinery, something that isn’t guaranteed in E. coli.
Does everyone understand this part? If you do plasposon mutagenesis as part of your project you will tkae a similar approach to “rescue” the mutated region of genomic DNA..
The interesting thing in this section is that they find a number of pseudogenes that are truncated, suggesting the A4 genome is pretty dynamic and that rearrangements sometimes lead to formation of these truncated genes that are missing regions that are necessary for activity.
They can’t show all the data and this information is not critical to the “story” they’re telling in this paper. You don’t have to show all the data. Sometimes editors make you eliminate some figures because they’re not essential.
What you should focus on here is understanding which clones can form indigo and which can’t and why. What genes are essential for indigo production from indigo?
It allows you to understand the regulation of the acenaphthene degradation genes. They can see where RNA polymerase biinds. They can examine that region for other regulatory binding motifs. They found an activator and it’s binding site but there could also be regulation by a repressor protein (think of the lac operon and how it is regulated by both an activator (CAP) and a repressor (LacI)).
It may help to recap basic PAH metabolism to understand how to iinterpret the mutant data. Think about the initial steps in PAH degradation (it may be helpful to review Figure 1 from paper 2). The first step is catalyzed by the dioxygenase enzyme (with help from ferredoxin and reductase) and converts the PAH to a diol by adding both molecule of molecular oxygen across a double bond in one of the rings (acenaphthene is converted to acenaphthene cis-1,2-dihydrodiol and to 1,8 acenaphthenol by a subsequent step). This is the dioxygenase can also convert indole to indigo.
Based on this knowledge a mutant that can no longer convert indole to indigo must be mutated in a limited number of genes. Make a list of all the possible genes that could lead to an indigo-minus phenotype.
After the initial dioxygenase steps there are a series of reactions (~5-7 steps) before the PAH is converted to a central metabolic intermediate that can be oxidized to yield ATP/NADH/NADPH to fuel growth. Mutations in any of these later steps would be able to produce indigo but would not be able to grow with PAH as the sole source of carbon and energy.
Inducers are effector molecules that bind a regulatory protein to alter it’s activity. to alter arhR expression we’d need to look at the regulatory mechanism that contrls transcription of arhR.
One possible explanation for the constitutive expression of arhR is that there has been some mutation in the arhR regulatory regions (promoter/operator) that have lead to continual expression of arhR.
Or, it could be that arhR is continually expressed by design. Remember that the lac operon is continiually expressed at a low level (necessary so that when lactose is present LacZ can convert it to the induce allolactose) but it’s transcription increases 1000-fold when the inducer allolactose is present.
I would say that it’s still worthy of study – after all it works! In class we learn about models like the lac operon that are often quite simple (and efficient) but don’t always represent what happens in non-model systems. What we see in A4 is that the genome is dynamic and constantly being remodeled by transposition etc. Sometimes this is advantageous, for example when A4 acquires useful genes such as acenaphthene degradation genes. Other times it can be deleteroius, such as when the transposition event yields pseudogens that lack essential prts of the gene (such as in ORF11).
I think it’s likely that other regulatory elements may come into play. This is a catabolic operon and may be subject to both positive control (by ArhR) and negative control (by some as yet unknown regulatory protein, maybe a repressor-type).
It’s mostly a lack of studies, although in my opinion the authors are overstating this “lack.” There are quite a few studies on biodegradation of PAHs by marine bacteria and the pathwyas to date are quite similar no matter where the bacteria were isolated (terrestrial vs. marine).
This is a reasonable assumption. If you look at the response of the Gulf of Mexico bacteria to the Deepwater horizon spill it was clear that the bacteria were already primed to degrade the oil from both years of small scale spills and the existence of many natural oil seeps in the Gulf.
I think the goal is more to understand the mechanisms/pathways for PAh biodegradation rather than to develop commercial super-degraders, although there are some commercial strains on the market for biodegradation of chlorinated hydrocarbons. The chlorinated compounds are much more persistent than PAHs so it makes more sense to add specialized degrading bacteria to those spill sites. PAHs are degraded on much shorter time scales naturally and the most you can do to enhance degraration is to add nutrients (N-P-K, like for the plants in your garden) and maybe a co-metabolite (easier to degrade carbon source to get the bacteria started).
I believe that the LB126 paper may have alluded to this. These are all mono-aromatic rings that are central intermediates in the degradation of a variety of PAHs. These intermediates are catabolized by a “lower” pathway that converts them into TCA cycle intermediates. The “upper” pathways are more specific to the PAH (e.g. phenanthrene vs naphthalene vs fuloranthene) whereas the “lower” pathways are more universal.
The cost is down to about $250 for a bacterial genome so not too pricey for a well funded lab. The advantage of doing the full genome sequence is that you’re not biased by previous research. You can (in theory) find novel genes that haven’t previously been described (especially if you use techniques like microarrays or proteomics).
GIs are regions of gDNA that look like they were acquired from an unrelated organism. They are often identified by looking at %GC as this % is usually the same value throughout a bacterial genome. If you see a region of gDNA with a markedly different %GC it was probably acquired by HGT.
In the next section they’ll also present all the potential PAH degradation genes they identified though sequence homology. This is essentially a list of potential targets for directed mutagenesis.
We’ve been reading about “oddball” sphingomonads which undergo a lot of genomic rearrangements. It’s more typical to have all of the PAH degradation genes next to one another.
It’s hard to say with out looking at the entire genome sequence. It may be that C. indicus has pathways for synthesis if all the amino acids while B30 doesn’t. It may be that C. indicus evolved in an environemnt where PAHs were abundant and that over time it has lost amino acid transport & metabolism genes (relative to B30) because the supply of PAH was so high that it didn’t need to metabolize amino acids.
Or, the differences in gene content could simply be random and have no effect on the relative fitness of the two strains.
They don’t tell us where the genes are (chromosome vs plasmid) yet. We’ll have to read on. The big thing to notice here is that there are a lot of genomic islands in the P73 genome. It seems to be a strain that is good at acquiring and integrating foreign DNA into it’s genome. It may have some of the DNA uptake proteins we discussed in BLY 314.
Yes, they’re talking about HGT here and HGT from different sources. There have likely been multiple events where P73 took up and incorporated foreign DNA – both plasmids and the genomic islands discussed earlier.
It’s possible that they were lst somehow, especially given that so many other chemotaxis and flagellar genes are present. They don’t tell us here if the che & mcp genes are in the core genome but they probably are.
They have the entire genome so they’re likely to find more genes just because they’re looking at way more DNA sequence. They’re also lucky in that the genes are located next to one another unlike in many of the Sphingomonas species we read about in our earlier papers.
If it still had a functional flagellum it could move towards teh source of PAHs. Remember how we looked at regulation of chemotaxis and flagellar motion in BLY 314?
These matabolites are produced during metabolism of a variety of PAHs. For example, some bacteria metabolize naphthalene with phthalate as an intermediate, others proceed through a salicylate intermediate, while still others go through gentisate. It’s a bit strange that P73 shows genetic evidence for many different PAH degradation intermediates but given the amount of HGT it seems to have undergone maybe we shouldn’t be surprised. It may have acquired PAH degradation genes from a variety of sources, each of which used a different “lower” degradation pathway.
Think about the arrangement of the lac operon. What genes does it contain other than beta-galactosidase? Is one of the genes a lactose transporter gene? Why would it make sense to have the degradation and transport genes in one operon? under the control of one promoter?
I think I’d phrase it the other way around – that the ability to take up a wide variety of compounds helps P73 degrade those compounds – I’d consider transport/uptake the first step in degradation
They could, although they may not have seen such clear results. Given that they identified a number of candidates that would facilitate PAH uptake it’s likely that there is some degree of redundancy and that you wouldn’t see a clear loss of function the way you do when you knock out the gene that encodes the first step in the degradation pathway.
There are other techniques that they could assess whether genes are linked to growth on fluoranthene – can you suggest any?
They found putative PAH degradation genes in four regions of the genome and labeled them regions A, B, C, and D to make discussing their findings easier. It’s just an operational definition rather than a concept you should remember from BLY 302/314!
This is true. In many ways I think genome papers are boring. They tend to be very descriptive, reporting on a laundry list of genes and what they might do, with very little experimentation. This paper is actually pretty good in that they did some directed mutagenesis experiment to test some of their bioinformatics predictions.
It’s really the reverse, where the information from papers like our previous papers allow the computational predictions. We need papers where gene function has been verified experimentally before we can use sequence homology to predict function as they did in this paper.
The significance is that in contrast to other strains which seem to add the oxygens at multiple positions, P73 only utilizes one option (addition of oxygens at carbons 7 and 8). This should make elucidation of the subsequent steps in the pathway easier as there aren’t multiple degradation pathways being used at once.
Yes, because they have the entire genome they can identify all the potential PAH-related genes computationally and then map functions on them through trying to match metabolites to particular enzymes they detected computationally or by knocking out genes and seeing how the phenotype of the mutant differs from the wild type.
Contrast this to the approach you (and the first three papers) took where you had to spend a good deal of time simply finding a gene to test.
In some ways this paper is a preliminary study. They sequenced the genome and discovered a large number of potential PAH degradation genes. They did some metabolite analysis to map functions to some of the genes and did mutagenesis to verify function of another. However, a lot more work could be done (verify function of the other genes).
The study of all the other PAH genes they identified represents probably 3-4 more papers. We can’t expect them to do it all in one paper. For one thing, many journals have page limits for their articles. For another, this article already presents us with a lot of information (especially if you read the 129 page supplemental information!)
This is something you should look up. I’ll post the references so that you can check the references cited to support this claim. Also, try to focus questions on the microbiology aspect of the paper. While the harmful effects of naphthalene etc. on mitichondrial respiration is important we want to focus on the isolation and characterization of bacteria capable of degrading PAHs.
We’ll find out as we move through this paper how long the isolates in this study take to degrade naphthalene. It depends based on the composition of the contaminating material.
Who/what do you mean by “those”? Any organism with mitochondria could potentially be harmed if the dose was large enough. It’s usually the larger PAHs that are harmful, not naphthalene. Naphthalene is the active ingredient in mothballs which you can buy and WalMart and use to protect your clothes. It’s harmful if you ingest it but you’d have to inhale a really large dose to suffer harm.
The authors are referring to damage to eukaryotes in PAH-contaminated environments. You’re correct about the difference in location of respiration in bacteria and eukaryotes.
It depends on the bacterium and the particular PAH. If you calculate the energy yield from the oxidation of a molecule of naphthalene all the way to CO2 and compare it to the energy yield for the oxidation of a molecule of glucose all the way to CO2 you’ll see that naphthalene is a pretty good growth substrate.
Combustion is one possibility. You send the contaminated material to an incinerator. You can also do chemical oxidations such as the Fenton oxidation where iron ions are used as catalysts.
Bioremediation happened naturally in the Gulf of Mexico after the Deep Water Horizon blowout. Naturally occurring oil-degrading bacteria bloomed in response to the released oil and degraded a large fraction of the oil and gas that was released into the Gulf. Most of the time you don’t have to add bacteria, they’re already there. you might have to add some fertilizer because the oil has a lot of carbon but lacks trace nutrients (N, P, K etc).
Why do you transport or store perishable food items on ice? To slow down microbial activity. Why would they want to slow down the activity of their samples?
They’re making stock cultures that they can use in the future. Storing a culture in glycerol between -20 and -80C keeps the cells viable indefinitely. They can sit in the freezer for years. It means you don’t have to worry about transferring the cultures onto fresh media weekly to maintain viable stocks.
For the second part, the authors just told you they chose phenotypically different colonies, colonies that have different appearences. They don’t want to isolate the same species 20 times, they want different species.
We’ll talk about this in a few weeks and do PCR in lab later this semester. Basically PCR is a way to make many copies of a piece of DNA from a target. It uses 2 oligonucleotide primers and a thermostable DNA polymerase. Because you need single stranded DNA to carry out DNA synthesis you first melt (denature) the double stranded DNA at 94-100C. Next you lower the temperature so that the primers can bind (anneal) to the priming sites. Then you increase the temperature to allow the DNA polymerase to make copies of the DNA. You repeat this process 25-40 times to end up with millions of copies of your target. See these videos for more details: https://www.youtube.com/watch?v=nHi-3jP6Mvc or the video in the study area in Mastering Microbiology
The other main feature of ONR7a is that it contains all the major ions found in seawater. It’s synthetic seawater. It’s also a defined medium, not a complex medium. It’s also selective because limiting the carbon and energy source to naphthalene means that only naphthalene degraders should grow on it.
The MSM (minimal salts medium) we used in lab and supplemented with naphthalene crystals is similar in that it serves as a proxy for soil/freshwater systems.
Yes they’re using an enrichment technique. By limiting the carbon and energy source to naphthalene they’re selecting against all the bacteria that can’t use naphthalene. See my reply to Bently’s comment on the previous paragraph.
These tests just tell us more about the bacteria and let us know if we isolated 54 different strains or if some of the 54 are the same strain/species. Before DNA sequencing of the 16S rRNA gene became the standard for identifying bacteria you would do a variety of staining and biochemical and physiological tests and compare them to results in Bergey’s manual to identify an isolate to genus and species.
Think about the surface properties of the bacterial cell and how that might affect the interaction of the bacterial cell with naphthalene. Is naphthalene hydrophilic or hydrophobic?
Some bacteria may be adapted to growing on high concentrations of naphthalene while others are better at growing on low concentrations. This could have applications if you wanted to use some of these bacteria to clean up spills depending on the amount or concentration of oil or naphthalene spilled.
Biology is variation! Think about our class. We’re all members of the same species but have vastly different phenotypes. Why should the bacteria be any different?
Perhaps. They could also be autotrophs or they may have been growing on waste products released by other bacteria on the original isolation plates and not on the naphthalene or they could be autotrophs. There are a lot of possibilities.
According to the 16S sequencing they’re different genera and they probably have different surface properties that account for the different BATH scores.
I’m not sure what your question is. Do you mean the ancestor of all the strains isolated? Not all the species represented on the tree are naphthalene degraders.
Phenotype/function doesn’t really follow phylogeny in the bacteria, especially not for functions that can be carried on mobile genetic elements such as plasmids and transposons. A lot of genetic variation in the bacteria is transferred laterally rather than vertically. This means that the species a bacterial isolate belongs to doesn’t necessarity tell you how that isolate will behave.
That would be a flask with naphthalene and media but no bacteria. So any decrease in naphthalene concentration would be due to things like photooxidation, sublimination etc.
This is true and interesting. The naphthalene degradation enzymes, transporters etc. in each isolate clearly differ to yield such different degradation rates. The surface properties measured by the E26 and BATH tests are also a factor in the different degradation results.
Is naphthalene hydrophilic or hydrophobic? Would a more hydrophilic or hydrophobic cell surface “help” a bacterium attach to a naphthalene containing oil droplet and degrade the naphthalene?
It may be that above 400ppm the naphthalene begins to exert toxic effects on the cells. Up to 400ppm increasing naphthalene concentration promotes growth (more C,E, e- to fuel cell growth) but above 400ppm the harmful effects outweigh any benefit from the increased C/E/e- supplied by teh increased naphthalene.
Think back to organic chemistry. Why are aromatic rings so stable and why would compounds with more rings be more stable?
It’s a bit more complicated than the number of rings as PAHs with the same number of rings can have different stability depending on the arrangement of the rings for example phenanthrene is more stable than anthracene even through they are both 3-ring PAHs.
Keep in mind that the teichoic acids are negatively charged. They probably wouldn’t enhance interaction with a hydrophobic substrate. Their function is to provide some rigidity to the cell wall and to bind cations such as Mg+.
The numbers are pretty constant for similar environments but the species composition will vary according to environmental conditions (nutrient rich vs nutrient poor, light availability, organic matter content etc.).
The Roling study didn’t isolate individual species, they examined the bacterial community that emerged in response to oil and identified them by sequencing 16S rRNA sequences obtained from the total community DNA, not from purified isolates. They didn’t directly link the genera identified in the community to oil/naphthalene degradation. This current study makes the link explicit by working with pure cultures.
You could use media that favor the growth of gram positives. You could also pasteurize the water sample. This would kill many gram negative bacteria and allow the gram positives to be more easily isolated, especially any endospore formers in your sample.
I think that one take home from this paper and the work we’re doing in the lab is that these naphthalene degrading bacteria are ubiquitous. If we use growth conditions that favor them (ONR7a or MSM + naphthalene) we can often isolate them. Thus, many environments are poised to respond to things like oil spills. We may not need to add any special bacteria.
It’s possible but it could also simply be a sampling artifact. If you were to search the literature you’d see that a wide variety of both Gram negative and Gram positive bacteria capable of PAH degradation have been isolated.
Remember the basic principles of evolutionary biology. The environment doesn’t cause anything. Mutations occur all the time in species. Some confer fitness advantages others are deleterious. In oil contaminated environments bacteria that had mutated to be able to utilize PAHs would be more competitive than those lacking that ability. The environment selects the fittest organisms but it doesn’t cause them. Mutations conferring the ability to degrade oil also occur in pristine environments but they probably wouldn’t persist in those environments because they wouldn’t confer a fitness advantage in that sitiuation.
The bacteria can degrade a lot of oil but some components such as asphaltenes and resins are resistant to biodegradation so there is always a need for physical oil removal as we saw in the Gulf of Mexico after the DWH blowout.
Also, in some environments you need to add fertilizer (mostly N and P) for maximal degradation. This is because the oil is carbon (and energy and electron) rich but nutrient (N/P?K etc) poor.
It could cause all the effects listed in the first sentence of your comment. In fact, if you cooked your seafood on your backyard grill you might add to the PAH load of your food!
This paper dates from the mid 2000’s when whole genome sequencing was still expensive and time consuming. Instead of sequencing a genome to find a gene you would usually fragment and organisms genome into smaller fragments and put it in a cloning vector (they use a phage system in this study) and transform the vectors into a host such as E. coli. The collection of E. coli clones was called a genomic library because it contained all the genes from the organism you were studying. It was easier to study the genes in E. coli than in the original organism.
Then all you had to do was find the E. coli clone that contained the genes you were interested in. How did they do that in this study?
Here they’re just describing the various techniques they used to manipulate and detect the DNA & protein they worked with in the lab as well as how they analysed the DNA sequences they obtained.
The dioxygenase enzyme needs a supply of electrons for activity. The ferredoxin and ferredoxin reductase ferry electrons from NADH or NADPH to the dioxygenase. There are many different ferredoxins that interact with a variety of enzymes in teh cell.
They’re not trying to biotransform the cells, they’re doing an experiment to see which aromatic compounds (listed in table 2) the cells containing pPhnA can biotransform.
Look at table 1. It lists the genotypes of the cells and plasmids used. The pPhnA plasmid contains Ap-r which encodes for ampicillin resistance. You add ampicillin to the growth media to ensure the E. coli cells maintain the plasmid and express the genes it contains (including the cloned dioxygenase gene).
What they know here is that they found two clones that share a common Sau3AI fragment. One probably has more DNA to the left while the other has more DNA to the right but they share a middle portion.
They also know that both of the clones contain the genes necessary for the first step in PAH degradation (and maybe more steps, we’ll find out below).
Because it is! The genes on a genome don’t all face in the same direction, although genes in an operon usually do, that way the proteins they encode can be translated off one polycistronic message.
The fact that orf7 faces in a different direction suggests that it may not function with it’s neighbours. It may be subject to different regulation. It may not eve be involved in PAH catabolism! The fact that it doesn’t have a “phn” name suggests it’s not involved.
Think about everything we learned about mobile genetic elements. These genes are often acquired through horizontal gene transfer on plasmids or transposons. Rearrangements can occur, especially during transposition. How would you look for transposon “footprints” in this region? You have the DNA sequence.
The identities are not bad for proteins! And the identities are probably much higher if we zoom into the catalytic and substrate binding domains. They wold probably find better matches today because the database is so much larger.
This paper was published in the early 2000’s. They’d probably find a better match if they redid the search today. You could perform it yourself and see! They gave us the accession numbers so we can pull up their DNA sequences from Genbank.
You need both subunits for activity. It may be that the small subunit is located elsewhere on he chromosome or that the PhnA1b is non functional. There could have been a gene duplication event that duplicated the large but not the small subunit gene. Given the conservation in the Rieske and substrate binding sites the first explanation is more likely.
Yes but it looks like it’s paired with a different large subunit, PhnA1. PhnA1b needs a PhnA2b small subunit. The gene naming takes a little getting used to and they’re a bit sloppy. PhnA1 should would more correctly be named PhnA1a.
The big thing to notice on the tree is that PhnA1 clusters with dioxygenases previously reported to initiate attach on PAHs while PhnA1b is very distantly related and clusters with dioxygenases that act much further down the degradation pathway.
Proteins with as little as 30% identity can catalyse the same reaction. The key is he identity at the catalytic and substrate binding sites. That’s why they zoom in on the Rieske and mononuclear iron sites in this paragraph.
I would say requires, not allows. It’s using the oxygen to break a C=C double bond in the PAHs. It’s not using it as a terminal electron acceptor in respiration in this case. These PAH degraders need oxygen for both processes. The dioxygenase pathway can only be used if oxygen is available.
I’m not exactly sure what you’re asking here. 4 genes are necessary for activity: A1 & A2 for he dioxygenase and A3 and A4 to transfer electrons to the dioxygenase so that it can add two oxygen molecules across the C=C double bond and break the aromaticity.
What did you learn about meta vs paras ubstitued rings in Organic chemistry? Electron donating vs electron withdrawing groups? Maybe someone else can help us out. You’ve covered this material much more recently than me!
Would it be better to make a specific ferredoxin and reductase pair for every dioxygenase the cell encodes or to have one pair that can partner with many different dioxygenases? Which approach would better conserve resources?
If the electron transfer proteins can partner with many different dioxygenases it would make sense for them to be regulated differently. THey’d meed to be controlled by multiple inducers, not just the inducer(s) controlliing transcription of phnA1phnA2.
Basically whether it has an enzyme that can oxidise arsentite (releasing electrons that the microbe can use for ???) or reduce arsenate (for example as a respiratory terminal electron acceptor).
Think about why the microbes are using arsenate in particular. Would they still use it under oxic conditions or would they have more energetically favorable alternatives available to them?
What is the electron donor in chemoheterotrophy? What are the possible electron acceptors (other than oxygen)? You need to think about which As species is oxidized and can act as an electron acceptor and which is reduced and can act as an electron donor.
There are plenty of planktonic arsenic utilizing bacteria. Here the biofilm lifestyle may allow aerobic and anaerobic arsenic utilizers to grow in close association and cycle AsIII and AsV between them.
Yes, but we’ll see anoxygenic phototrophs in this study so it will be photosynthesis with a twist. One of the differences is that the phototrophs will use something other than water as an electron donor during photosynthetic electron transport.
This is something you could look up in a scientific database such as sciencedirect, PubMed, or Scopus and report back to us. Jest search for arsenic resistant bacteria.
Many arsenic resistant bacteria exist but not all can use AsIII and AsV the way the bacteria in this study do.
It’s just a physical gradient – grater distance from the source of the hot water with the resultinig temperature selecting different bacteria and/or archaea.
Exactly. They want to preserve the communities in the state they were in when they samples so they lower the temperature to slow biological activity down and put them in the dark to halt photosynthesis.
They don’t want to kill the samples as they want to use some in future incubation experiments so they store at 5C rather than freeze them.
They probably can’t. More importantly N2 isn’t usually used as a terminal electron acceptor in ETCs (why not?). By saturating the bottles with N2 they eliminate aerobic respiration and keep the samples in a state close to their natural habitat before using in their experiments. It should keep the community very similar to it’s original structure and activity.
Shaking the aerobic tubes/bottles increases aeration of the cultures and ensures that oxygen doesn’t become limiting at the bottom of the tubes/bottles.
Here they are referring to libraries made by ligating an PCR product into a vector. You could also make a genomic library where you ligated fragments of genomic DNA into a vector (what awas done in our previous discussion paper).
It allows them to add a very small amount of radio isotope and track where it goes. It means they can leave the natural concentrations of AsII and V virtually unchanged.
There’s no antigen here, antigens react with antibodies, but you’re on the right track. The use of isotopes makes it easier to measure what is happening to the arsenic or the carbon radioisotopes that they add to the samples. A small amount of isotope gives a large easily measured signal so they can add a little isotope and track what happens to the C or As.
The dominant clone was highly related to a bug they previously isolated from this same location. They also tell us that this bug, Ectothiorhodospira, strain PHS-1, can carry out some of the same processes they were exploring in the experiments in this paper. This is an example of a case where molecular biology approaches and culture based approaches give the same answer about which bacteria are dominating a system. This doesn’t always happen.
Yes. They include a killed control in order to show that any changes in arsenic speciation were caused by living cells, not just spontaneous chemical reactions.
They set them up that way for ease of analysis although not all the experiments started with only one member of the AsV/AsIII redox couple in the incubation. Once the incubations are underway we have a mixture of AsIII and AsV. Look at the 50h time point in figure 1 for example.
They ran it long enough to see all the AsIII oxidized to AsV and then for long enough to see all the AsV reduced back to AsIII. The biology of the biofilms dictated the length of the experiments.
Yes, but what you need to do is identify what kind of metabolic processes could account for these observations. During what kind of metabolism(s) would a cell oxidize AsIII (arsenite)? reduce arsenate (AsV)?
What processes would drive AsIII oxidation under oxic conditions? If the rate is the same with and without oxygen do you think oxygen is involved in the process?
Hard to say. It could simply be that the bugs using the arsenite are increasing in numbers as the incubation progresses and therefore the AsIII is oxidized more rapidly.
No, I think we’re just seeing a little bit of abiotic AsV reduction or just analytical error. The error bars on the measurements are pretty large. The AsV time zero and final time point for the no electron donor trace may not be significantly different from one another.
Think about “who” would use the acetate or bicarbonate and how they would use it. Remember, bicarbonate is essentially CO2 and will be processed by the Calvin cycle or some other carbon fixation pathway. Also remember that anoxygenic phototrophs can use a wide variety of electron donors and that microbes are versatile and may use a number of different metabolic strategies depending on environmental conditions.
What they mean by “not clear” is that they can’t tell which explanation is correct.
Can you think of a genomics or mutagenesis based experiment they could do to identify the arsentie oxidase gene(s)? This would be a great exam question.
We’ll discuss some techniques in class on Tuesday that should give you some ideas on how to link particular functions to particular microbes in a communtiy.
The previous paragraph tells us that acetate was not used as an electron donor (and carbon and energy course). If the acetate was used as a C, e- and energy source in anaerobic conditions it could use AsV as the terminal electron acceptor (ie stimulate AsV reduction). But they did not see AsV resuction stimulated when they added acetate in the dark.
But, in the light they see acetate oxidation so in the light some process that removes electrons from acetate . . .
You’ll see as the semester goes on that a lot of work has been done with naphthalene (2 rings) but not as much with the higher MW PAHs. Naphthalene is easier to work with (higher solubility) and much less toxic than the higher MW PAHs which is why we’re using it for our studies.
One thing to think about is whether there is any reason fluoranthene degradation would differ in marine vs. terrestrial bacteria. My feeling is the bacterial species may differ but that the biochemical degradation pathways should be very similar. We’ll find out.
True, oxygen availability will influence biodegradation but if we’re looking at aerobic marine vs aerobic terrestrial environments the pathways will probably be similar, although the microorganisms may be different.
Given that PAH degradation genes have been isolated from other microorganisms they use bioinformatic analyses to search for similar genes in the C. indicus genome once they sequence it.
This bioinformatic approach will only work if the C. indicus genes are similar to previously described genes. If C. indicus has a completely novel PAH degradation pathway they will need a different approach.
The previous paragraph describes annotating the C. indicus genome (inicluding, based on the last paragraph of the introduction section, PAH degradation genes). Given their research goals we can infer that they wanted to mutate (knock out) a PAH degradation gene using the cre-lox system.
It makes it easier to separate and identify the various intermediates in the fluoranthene degradation pathway. If we can identify all the intermediates we can figure out what enzymes are required for each step in the degradation pathway.
They probably used an enrichment culture technique where they put some sediment and buffer in a flask and added PAH as a sole carbon and energy source. That way, only PAH degraders would grow up. That is what the BLY 314 students did last semester to isolate the naphthalene degraders we’re working with this semester.
Exactly. If they knock out or inactivate a gene and the mutated bacterium can no longer utilize fluoranthene that would support the hypothesis that the knocked out gene is necessary for fluoranthene utilization.
The key s to read the figure title carefully and then look at the big picture for each ring. For example, we are interested in PAH degradation genes – probablt categorized as energy productin and metabolism, so we could focus on those colors.
We also know that non-essential (but useful) genes like PAH degradation genes are often acquired by horizontal gene transfer so we might also want to look at the %G+C and GC skew figures and see if there is evidence of HGT from te %G+C across the genome. Remember that organisms typically have a fairly constant %G+C and large deviations from that % are usually indicative of HGT events.
I disagree. Looking at the circles that depict G+C content, is G+C content consistent across the chromosome? across the individual plasmids? Given that G+C content is usually fairly constant across a bacterial chromosome or plasmid and that in these figures G+C content is not constant what can we conclude about the evolution of the genome of C. indicus? Based on the figures do you think the genome we see “today” is the result of only vertical evolution or has horizontal gene transfer also played a role?
Yes. This figure shows us that the RepA ad ParA proteins from the C. indicus chromosome and plasmids do not share a common evolutionary history. If they did they would all cluster together on the same branch of each tree. The trees are another line of evidence that the C. indicus genome has been remodeled by extensive horizontal gene transfer.
Given that the proteins from the various plasmids are related to a variety of different bacteria we can conclude that the plasmids all have different evolutionary histories and were probably acquired in separate events.
They’re closely related but one degrades PAHs and the others doesn’t. The comparison is analogous to comparing the genomes of two E. coli strains, one virulent, the other non virulent. It’s a way to identify all the genes that may be involved in PAH degradation, not just the ones that are similar in sequence to previously characterized genes.
You’d expect genes that were acquired from the same source to share a high degree of sequence similarity but also exhibit conserved gene order. The gene order can change over time if the organism you’re studying undergoes frequent genome rearrangements. All they’re doing here is showing us the degree of conservation of gene order between C. indicus and the two bacteria containing the PAH degradation genes with the highest degree of sequence identity.
Some transposons have sequence preferences for the sites they will transpose into, others are less specific. I think they focus on B here because the thrust of the paper is identifying the PAH degradation genes. It may also be that they didn’t identify insertion hotspots in the other regions.
You’d want to look at RNA, not gDNA. You could also use Reverse transcription PCR to examine when the gene transcript is produced and link transcription of the RHD gene to the presence of different inducers/substrates.
You could do qRT-PCR to quantify expression.
You could also make an expression clone that contained the RHD genes and transform it into E. coli and see if E. coli gained the ability to metabolize fluoranthene.
It’s because the aromatic structure is really stable. Even though we often draw the benzene rings with discrete double bonds the pi electrons are shared between all 6 C in the ring and attacking the aromatic ring is therefore more difficult than attacking a C=C double bond in an alkene.
Based on the annotations many of them do. Have a look at the degradation pathway figure. They’ve put the “code names/numbers” of the genes they identified above the various reactions. If you look closely, some of those steps are oxygenations.
Generally bioremediation for things like PAHs relies on naturally occurring microbes. We don’t usually add designer microbes to a PAH polluted environment. In contract, environments contaminated with chlorinated hydrocarbons do have “superbugs” added to them as the ability to attack chlorinated pollutants is not as common.
For PAH contamination we might add nutrients/fertilizers (N/P/K) to add the elements that are missing in the PAHs but necessary for biosynthesis.
Until this paper was published all the fluorene degraders that had been reported in the literature were Gram positive which was a bit odd. There is no biochemical or physiological reason that this ability would be restricted to Gram positives. After all, Gram negative bacteria degrade many other PAHs.
The biggest different is the ability to grow in 3.5% salt for marine bacteria. Other than that terrestrial and marine bacteria share the same biochemical pathways for the most part. Both environments contain a lot of proteobacteria.
Meta-cleavage refers to the position on the aromatic ring at which it is cleaved. In IUPAC nomenclature meta is at carbon 3, ortho at carbon-2 and para at carbon-4. See more on meta-cleavage here https://metacyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-5415
Note how the pathway ends with metabolites that can enter the TCA cycle, a common pathway found in most microbes.
I believe that GSL stands for GlycoSphingoLipid and LPS is liposaccharide (found in the outer layer of the outer membrane of Gram-negative bacteria) The latter you should remember from BLY 314!
Sometimes the initial dioxygenase has a much broader substrate range than the enzymes further down the pathway so you end up with situations where you have an RHD that can hydroxylate a variety of compounds but the products it makes cannot be processed by the enzymes catalyzing the subsequent degradation steps. For example they might not fit into the active site of the next enzyme.
Exactly. They included restriction sites at either end of the PCR primers so that they could clone the RHD into an expression vector in the correct orientation. Why is that important?
Gene knockout is definitely a possibility as well. Usually a reviewer will want to see two separate experimental lines of evidence that a gene is involved in a particular process. Expression cloning and knockout are two possibilities. This papers utilizes a third approach (RT-PCR) to confirm the expression clone results.
Remember that the purpose of GC-MS is to separate and identify metabolites. The sample is heated up and compounds with different evaporation temperatures will separate at different points along the temperature range allowing for their detection.
Yes. It makes cloning the RHD fragment easier by reducing the number of clones they have to screen to find the RHD containing fragment. It also means they should only get white colonies as the vector should not be able to self ligate because the sticky ends from the two different enzymes don’t match.
You’re on the right track. The sonnication breaks the cells open releasing the cytoplasmic contents. They don’t know if the RHD proteins are transported out of the cell or not so they lyse the cells to be sure that they “see” everything on the gel.
Think about all the information they gave us. LB126 can grow on fluorene (as it’s sole C source) but it can degrade phenanthrene, anthracene, and fluoranthene as long as you also supply pyruvate as a C source. This means that LB126 needs a supplemental C source when grown on the other PAHs because it cannot make all the biomolecules it needs for growth from them.
Think back to when you studies metabolism (both catabolism and anabolism). What do cells use pyruvate for? What pathways is it part of?
Previous work had found these lower pathway genes in LB126 (reference 47) and they were most similar to genes from Sphingomonas KA1 and CB3. They’re pointing out that these new genes also have some similarity to Sphingomonas genes but the matches are not as good as to the Gram positive genes. Maybe the other sphingomonads also acquired genes from Gram positives???
Look at the yield column in the table. Substrates that yield a high % of product are “good” substrates. Does fluorene yield the highest % product? What product(s) are produced from fluorene? Do we see the expected product? Was there anything unexpected here?
Pyruvate can also serve as a precursor molecule for many important biomolecules. When they incubate LB126 with the non-fluorene PAHs they need to supply a supplemental C source that can be used for both energy generation and biosynthesis as they can’t metabolize the other PAHs all the way to all the intermediates they need for those purposes.
No. It just means that E. coli was making the proteins but not folding them correctly so they couldn’t attack fluorene. They had to change the growth conditions (grow at 42C instead of 37C) to get more of the protein make in the active form.
Maybe, , but maybe not. The structure of the enzyme is also important. For example you could have an RHD whose active site fits fluorene better than naphthalene and therefore degrades fluorene more efficiently.
It has to do with which carbons on the PAH are attacked by the RHD. Naphthalene, for example is attacked at the 1,2 positions by “lateral” dioxygenation. Dibenzofuran is attacked at the 4, 4a carbons in an “angular” dioxygenation. See this figure
It’s hard to say as there is not as much work on acenaphthene and acenaphthylene biodegradationas on PAHs like naphthalene and phenanthrene. However, we’ve seen similar patterns in the previous two papers where one PAH can serve as a sole C ad energy source but other PAHs cannot (even when the RHD can attack other PAHs). It will be interesting to see if the RHD these investigators identify follows a similar pattern.
They’re not exactly the same but they’re closely related. After all, in the last paper we read they used degenerate PCR primers that were able to amplify RHD genes from a broad array of bacteria.
That’s right. Molly has more details above. They used it anytime they needed to cut a band out of an agarose gel and purify it so that they could clone it. For example you could digest genomic DNA, run the digest out on a gel, cut the size range you wanted out of a gel, and ligate it to a vector cut with the same enzyme.
They found two copies of the arhA1 gene (probably saw two different bands on a gel in a Southern blot with an arhA1 probe). So now they have to wonder if both copies are functional/active or if one is a pseudogene.
Drawing a radial tree like this one is really just a matter of preference. Some people prefer this layout over the “boxy” trees we drew. MEGA lets you do either.
Ideally they’d clone and sequence the homologue so that they could figure out why it doesn’t confer dioxygenase activity. It could be a partial gene that has enough sequence to bind the Southern blot probe but is missing sequence necessary for dioxygenase activity.
It means that it’s related but not closely related which makes sense as acenaphthene and acenaphthylene are different enough from naphthalene and other PAHs that the acenaphthene dioxygenases could be expected to differ from the Nah-like RHDs.
Putting the RHD on one plasmid and the ferredoxin and reductase on a second plasmid means they could test a variety of RHDs in a host that contained the ferredoxin+reductase plasmid by just adding a second plasmid that contained the RHD genes. These genes are not always next to one another so making a clone that had RHD-alpha+RHD-beta+ferredoxin+reductase on one plasmids would be more difficult than using the two plasmid system.
You could use RT-PCR to detect expression of transcriptional regulators but more often when studying regulators you fuse the regulator promoter to a reporter gene and measure the activity of the reporter gene.
The gene clusters have promoters that are all switched on/off by the same effector molecule. Remember in BLY 314 when we studies regulation we compared the lac operon to the mal operon. In teh lac operon the catabolic genes are located together but in the mal operon they’re in different locations on teh chromosome but in both cases they were controlled in the same general manner (negative control of an inducible gene – repressor being inactivated by binding of an inducer molecule).
Today we’d probably sequence the entire genome. When this paper was published that would have been very expensive, hence they take a more limited approach.
They’re just pointing out a limit of their previous study and hinting that the found those genes this time. It could be that the ARhA1A2 protein will have even better activity with its true electron transfer partners.
If you’re on campus you can access the journal. At home remote login to the library to see the tables. I’ve fixed the links so you can see the tables here now.
They did a lot of subcloning using restriction enzymes. The TA cloning we did is more efficient (and cheaper to do than when this paper was published).
It “marks” the insertion – Gm resistant colonies are mutated, Gm sensitive colonies are not. It also allows them to clone out the mutated region using the Gm resistance as a selectable marker.
It helps here to remiond yourself what the first few steps in acenaphthene degradation are. 1,8NA is a downstream metabolite of acenaphthene. You could lose the ability to carry out step one ion the degradation pathway but still be able to do steps 2-10 if provided the substrate for steps 2-10 dpending on where the mutation occurs. Is it in the dioxygenase genes (disrupt step one but still have the ability to carry out steps 2-10) or is it in a regulator (lose ability to carry out any and all steps in the pathway because no mRNA is transcribed for any genes in the pathway).
These are all good questions. generally we’re using bacteria that are “native” to the contaminated site or from a similar site so they don’t disturb the ecosystem. In terms of downsides bioremediation can be slow and can be limited by other nutrients such as N and P. This can be overcome by adding fertilizer to the oil spill area so that the oil provides the carbon for the bacteria to grow (and respire the oil to CO2)and the fertilizer adds the other required nutrients for biosynthesis of new cells.
Think back to what you learned about aromatic and polyaromatic hydrocarbons in organic chemistry. Which do you think would be easier to break down, a 2-ring or a 4-ring PAH?
Some of the oil ends up as bacterial biomass and some ends up being respired to CO2. The ratio depends on the growth efficiency of the bacteria. Basically you see a transient bloom of bacteria that is then grazed by larger microbes so the oil carbon moves up through the food web with more and more being “lost” as CO2 at each trophic transfer.
They’re not changing the concentration of bacteria in the environment in this paragraph. They’re just reporting where the obtained their samples and how they transported them.
Why do you store anything on ice or in a fridge or freezer? To slow down or prevent microbial growth. They want to preserve the sample “as-is” until they get back to the lab.
Focus on the experimental methods in your comments on this section. I want to make sure you understand the approach the authors took in their quest to achieve the goals outlined in the introduction.
I’m not sure I agree with you on this. What are their experimental objectives? In this section they’re isolating NP-degraders so they’re not concerned with replication or repeatability. They simply want to isolate as many different NP degraders as possible.
They’re measuring the density of the bacteria in the culture by measuring the OD (see the microbial growth chapter in our text). They’re not measuring DNA. For that you’d need to measure at much shorter wavelengths.
At the beginning they worked with 50+ isolates. This first method is faster and easier and accurate “enough” to allow them to distinguish between slow and fast NP degraders.
What do you think they should measure instead of the amount of naphthalene left (i.e. not metabolised)? Measuring the amount of NP left in the culture vessel is straight forward and gives you the amount used by the bacteria by simply subtracting it from the amount added at the beginning of the experiment.
There are lost of analytical methods to choose from. Our focus should be on why they made the measurement, not on how.
They have 50-something isolates. How will they figure out if they are all different or if some of them are different species? By doing morphological (staining), biochemical & physiological, and molecular (16S sequence) tests on them an d comparing the results.
What do you know about the bacterial cell surface (hydrophilic or hydrophobic)? PAHs (hydrophilic or hydrophobic)? What kind of cell surface would attract PAHs?
The shaker allows for better air exchange so that the media remains oxygenated. What do NP-degraders need for the first step in aerobic NP degradation (I put this on the board in lab).
This is like asking why your brother is taller than you are! If we had the time to really dive into this question we could look at things like differences in the gene sequences coding for NP uptake and degradation but lets just say they’re a lot of natural variation among bacteria.
It keeps the cultures aerated. Remember thet first step in bacterial degradation of NP that I put on the board in lab? What was the other reactant in that first step?
You assign your isolate to a genus and species by comparing it to previously described species and based on the level of identity you assign your unknown bacterium to a genus and or species. For example, for bacteria, if the %identity of the 16S rRNA gene is greater than 97% we usually classify them as belonging to the same species.
If they only put their isolate sequences in the tree we’d be able to tell how closely related they were to each other, but we also want to include reference sequences so that we can compare them to the larger bacterial family.
Both the E24 and BATH tests measure how well the bacteria can attach to the hydrocarbons. The bacteria need to be able to “touch” the PAH to degrade it but most bacteria have hydrophilic surfaces and PAHs are hydrophobic so PAH degraders need to modify their cell surfaces.
As mentioned below it could be toxicity and probably is. If the bacterial transport or enzyme systems were saturated we’d expect to see a plateau, not a decline.
Look at the % NP degraded after 15 days. For some isolates it’s almost 90%. if they incubated them much longer there would be nothing for them to grow on.
No. It is just natural variation. Just look at our BLY 314 class, we’re all members of the same species but we show a lot of variation in our traits. Why should the bacteria be different. The isolates in this study are more diverse than our class, with a number of different genera present so variation in the rate at which they degrade NP and their BATH and E24 results are to be expected.
This is a tricky question as you have t decide how to measure the concentration – what volume of seawater are you going to assess? The concentration at the sea surface or throughout the entire water column?
The more rings, the harder the PAHs are to degrade. Naphthalene (2) and phenanthrene (3) are degraded pretty rapidly, pyrene, chrysene, and benz[a]pyrene more slowly.
If you do a search you’ll find a lot of papers describing naphthalene (2 rings) and phenanthrene (3) degraders but much fewer reports of pyrene(4) or chrysene (4) degraders and hardly any on 5-rings compounds like benz[a]pyrene.
You’ll also find a lot of reports on linear alkane degradation but few if any on branched or cyclic alkanes.
Some genera are found all over the place others can show some geographic distribution patterns. There is no hard and fast rule. Some like to quote Baas-Becking: “Everything is everywhere, but the environment selects.”
The bacteria were already there but the addition of oil or NP essentially creates an environment where they have an advantage and become more abundant. We can think of an oil spill as an ecosystem scale enrichment experiment. Also, many of these PAH degradation pathways are carried on plasmids that can be transferred from one bacterium to another (across species, genus, and higher) so some of the isolates could have recently (in an evolutionary sense) acquired the ability to use NP.
NP is often used as a model because it’s safer to use in the lab whereas the 4 & 5 ring PAHs are quite toxic. We’ll see in our next paper that bacteria that can degrade NP can often degrade other PAHs albeit more slowly.
I wouldn’t consider it the main question so much as a slightly unexpected result. Both Gram negative and Gram positive bacteria have been shown to degrade NP. It may be that something about their experiment or the area they sampled favored Gram negative bacteria. I mentioned elsewhere that in the BLY 314 lab we typically isolate way more Gram negatives, usually because they grow faster.
Yes there are. Their numbers rose after the DWH spill but are back to normal levels now that the oil is either gone of buried in the sediments or converted to tar balls which are really resistant to biodegradation.
There are some examples of doing this kind of genetic engineering although not usually for PAH degradation as so many “wild” bacteria have the ability. Some enzymes from PAH degradation pathways have been engineered into E.coli to produce indigo and other commercially valuable molecules.
How do we usually construct phylogenies? look at evolution of genes and genomes? By looking at vertical transmission (parent to offspring). So adding in possible horizontal transmission of genes (between totally unrelated species) will introduce all kinds of confusion into the phylogenies of these genes?
How would you determine whether a gene has been subject to horizontal gene transfer (HGT)? One way is to compare the phylogeny of the gene of interest (C23O in this case) to the phylogeny of a gene that doesn’y undergo HGT (like the 16S). You’d expect the a gene that undergoes HGT to produce a tree that differs from the 16S tree.
Right. You’re doing a scaled-up version of what we did in lab last week, a bunch of tests in a microtiter plate instead of in test tubes. See here for more info and pictures: Biolog .
Look at the numbers in the primer names. They’re not amplifying the entire 16S gene but they’re getting most of it so it will be enough to assign the isolate a genus and most likely a species.
With respect to the database they used, different people have different preferences. The choice doesn’t usually affect the result.
Think about our discussion in class of how to determine bacterial abundance. Why would they dilute the cells to a particular turbidity (=OD) before adding the cells to the test plate.
They’re describing the construction of a genomic library here (see https://upload.wikimedia.org/wikipedia/commons/7/7e/Genomic_Library_Construction.png). Basically you cut the ZX4 genome into little pieces and ligate them into a plasmid and put the resulting plasmids into E. coli. Each E. coli gets a different piece of the ZX4 genome. E. coli can’t use PAHs so it’s a good way to find genes that play a part in PAH degradation. You look for E’ coli colonies that have gained the ability to metabolize PAHs or their degradation intermediates (like catechol).
The authors looked for meta-cleavage gens by spraying the colonies with 2,30dihydroxy biphenyl diether. If they were looking for clones that contained a dioxygenase gene, what would they have done?
They’re lysing the cells to release the enzymes into the bulk solution so that they can access the various substrates they’re testing in these enzyme assays.
Think about the citrate test we did in lab last week. All of out lab NP degraders are aerobes and likely possess citric acid cycles but not everyone had a positive citrate test. Why?
You’ll have to wait for the results to get specific information but you can make some guesses at this point. It probably grew well on phenanthrene compared to their other isolates given that they chose it as the focus of their paper.
This qualifies as getting bogged down in details! Our concern is not with how the instrument they used works. We care about what they measured with the instrument and what that tells them about their isolate or clone.
Ask a new question or, if you’re really interested in how FIDs work look it up and share it with us.
Read the previous comment by me for the answer to your first question.
Use google to look up how a centrifuge works. That question counts as getting bogged down in details that aren’t that important to understanding the paper.
SalI is a restruction enzyme. It cuts DNA at a specific sequence. If you’re trying to clone a piece of DNA you can cut your plasmid and your insert DNA with a restriction enzyme to generate compatible ends that you can stick together with DNA ligase.
Brittney has it. They’re using the TCM to extract the phenanthrene from the aqueous sample. You probably did something similar at some point in organic lab using a separatory funnel.
Because they’re trying to identify phenanthrene degradation genes from ZX4. They made a library of ZX4 genes and put it into E. coli and looking for phenanthrene degradation activity. If E. coli could degrade phenanthrene all their clones would appear positive. They need to use a host that lacks the function they’re trying to find.
The focus in this paper is on describing one isolate in great depth. They probably had other phenanthrene degraders but this was the best or the most unique so they decided to focus on it.
Yes these are fairly standard tests. You can always find more tests to run but given that the focus of this study is phenanthrene degradation by ZX4, so the tests listed are sufficient.
Yes, they use GC. The first sentence of paragraph 3 of the M&M mentions it but it’s really easy to miss. It’s a very standard analysis so many papers will not give the methodological details in the M&M. Here is a link to one of the major companies that do these analyses http://www.midi-inc.com/pages/microbial_id.html
The number is the measurement of the turbidity or absorbance of the cells and substrate in each well of the microtitre plate. Would you expect a high or a low number if the substrate was consumed by the cells?
It doesn’t produce the enzymes that break down those substances. A similar situation to why we can’t live off cellulose: we don’t have the enzymes necessary.
What do you mean by fix? In biology to fix something is usually to reduce an inorganic molecule into a biomolecule using ATP and NADH/NADPH (ex. carbon fixation and N-fixation).
When we’re looking at phenanthrene (and other PAHs) degradation we’re looking at an oxidative pathway that yields ATP and NADH for the cell. As with any organic substrate it could be respired completely to CO2 or it could be partially oxidized to TCA cycle intermediates which are used in biosynthetic pathways.
Did anyone compare these concentrations to the naphthalene concentrations tested in our previous paper? This would be a great comparison to include in your summaries of this paper.
No it is not. You need to think about this in terms of how catabolic pathways are regulated. Instead of glucose vs lactose, thin about glucose vs phenanthrene.
Catechol is an intermediate in the phenanthrene degradation pathway. When should the cell make catechol 2,3 dioxygenase?
It may help to draw the figure 3 gene map out and place the information from this paragraph on it (promoter location, RBS locations) What does this suggest about which genes are transcribed together?
Yes. Look at the structures and think about what you learned about the effects of different functional groups on aromatic rings in organic chemistry (an O-Chem II topic, I believe).
There are a few things that can limit growth on PAHs. The first is the toxicity of the PAH. There may be a concentration at which toxic effects overwhelm the ability of the bacterium to degrade the PAH. The second possible limitation is that there is a concentration at which the PAH uptake and degradation enzymes can’t go any faster or biosynthesis of other cellular components can’t keep up so growth stalls.
Remember here that they’re just reporting the 16S sequence match. 98% identity means the same species but could be different subspecies or strains. The G+C% and the 16S sequence support each other in categorizing ZX4 as S. paucimobilis.
They picked 16S sequences from a variety of previously described PAH degraders. They build the matrix and then plot the data in tree form. The clusters in the tree are dictated by the matric/by the similarity of the sequences used to generate the matrix.
It depends on where the oil comes from. depending on the source crude oil has different percentages of aliphatic, aromatic, resin, and asphaltene type hydrocarbons.
These are two steps on the same overall pathway. In the upper pathway phenanthrene can be degraded all the way to salicylate. The salicylate can be converted to catechol or gentisate which are subsequently oxidized to 2-4 carbon molecules that can enter central metabolism (like TCA cycle) in what’s referred to as the lower pathway.
There are two major versions of the lower pathway: ortho cleavage and meta cleavage. They lead to slightly different end products because of where ring cleavage occurs.
I left out a lot of details but that’s the big picture. The authors are using the fact that ZX4 can utilise and transform certain intermediates to infer which of the possible lower pathways it uses.
There are reports of strains that only partially degrade phenanthrene and other PAHs. Sometimes it’s due to mutations in downstream genes that inactivate an enzyme in the degradation pathway. Othertimes a bacterium may not acquire the entire degradation pathway during horizontal gene transfer.
They told you about the meta-cleavage pathway in the results section, reread paragraph 8. It’s a little bit indirect but I think you should be able to figure it out. This is also something you could google quite easily.
They’re just saying that the ZX4 GST is more similar to EPA505 and F199 and less similar to SYK-6 and RW5. The sequence groupings match the CDNB activity. Presumably the differences in the DNA sequences lead to differences in amino acid sequence that lead to this difference in activity. It’s not a crucial finding for the paper but it is interesting.
They didn’t find the full GST sequence or gene. That’s a cloning issue, not a result of mutation. We’ll talk about it in lab tomorrow.
I think it’s a question of breadth vs depth. They’re both valid, thorough studies. They just have different goals: paper 1 wanted to assess/describe the PAH degradation potential of bacteria from the Persian Gulf. This study focuses on one strain and tries to understand how it does it (identify intermediates in the pathway, find the genes that code for degradation).
Do you know how indigo dye is normally produced? It might be that producing it using biotechnology would be more environmentally friendly or cheaper than how it’s currently produced.
I think the focus of a lot of these papers and of a lot of bioremediation is to understand which bacteria can bioremediate and how they do it rather than growing up batches or bioremediators and releasing them into the environment. The safety issue is always a concern for regulators.
The location of the promoter, coupled with the distance between phnH & I suggests that they are co-transcribed. The fact that phnG is upstream of the promoter suggests that is is transcribed (and regulated) independently of phnH & I.
Keep in mind also that organisms can use more than one nutritional strategy. One could be both a chemoorganoheterotroph (energy, electrons and C coming from organic sources) AND a photoorganoheterotroph. Many microbes can switch between various C, energy and electron sources depending on the environmental conditions in which they find themselves.
To answer this you need to look at the nitrogen transformations that occur during nitrification and denitrification. The last sentence may help you out.
It’s hard to compare without more information. For example we’d need to know what the electron donor was when we use AsV as the electron acceptor so that we could guess at the energy yield. We’d need to know what electron acceptor was used in the oxidation of AsIII.
I think it’s partly because arsenic is relatively abundant in the Mono lake system. I’m not sure that the other toxic elements you found are equally abundant in Mono lake.
One could imagine using AsIII oxidation to AsV as a detoxification mechanism. The key would be to ensure that there were no microbes present that could convert (reduce) the AsV back to AsIII.
Exactly. We can often see these kind of cooperative interactions across oxic/anoxic transitions or in environments that switch between oxic and anoxic conditions over time.
Under oxic conditions you’d never use AsV as an electron acceptor, O2 is much better. So if you want to see if AsV can be used as an electron acceptor you need to use anoxic conditions.
We’ll see that some of their experiments were run under both oxic and anoxic conditions but their focus is mostly on the anoxic conditions.
Think about what processes the AsIII and ASV could be used in. Some occur un the presence or absence of O2. Some will only make “energetic sense in one of he two conditions.
The green ponds were dominated by cyanobacteria and presumably oxygenic photosynthesis. My guess is that they were interested in studying other metabolic processes.
They’re keeping the slurries alive but slowing down their metabolism by storing at 5C. They froze the biofilms to immediately stop all biological activity so that when they extracted DNA and did PCR they would get a snapshot of the microbial community as it was a t the time of sampling.
They’re actually using N2 to create an anoxic atmosphere. It would be better to use argon as it is a noble gas and unreactive but N2 is cheaper so it’s often used instead.
With an immunoassay you have an antibody-based assay. In a radio assay you use a radioactive isotope of your analyte of interest as a tracer as it’s often easier to measure radioactivity that particular biological transformations.
For example, when measuring photosynthesis it’s easier to measure the incorporation of 14C-Co2 into biomass than to measure the actual sugars the CO2 was incorporated into. You add the radiolabeled CO2, incubate and then filter the cells out of the solution and measure the radioactivity level of the cells to measure photosynthetic CO2 fixation. The textbook discusses radioassays in chapter 19 (microbial ecology methods).
They were different, that is all. They used BlastX which takes a nucleotide sequence and translates it into all 6 possible reading frames and then compares those orfs to a protein database.
Think about the possible fates of the different radio-labeled substrates they use in this experiment. Why would they want to measure the amount of C-14 on a filter.
These incubations are closed systems. it’s likely that over time waste products are accumulating and growth rates are slowing down. Think about the bacterial growth curve for batch cultures.
I think you have a typo – arsenite can’t be both an electron donor and an acceptor. What are the electrons from arsenite donated to in anoxygenic photosynthesis?
We can’t really answer this “why” question. If you look at the 16S rRNA based tree of life you might conclude that the bacteria are more diverse but they’re less diverse than the Archaea in the red mat biofilm (but maybe not in the water or the green mat biofilm).
That’s reasonable. The other thing to focus onin this paragraph is how they explain the difference in AsIII oxidation rates in light vs dark incubations, suggesting that in the light both AsIII oxidation and AsV reduction could be occuring leading to the lower overall AsIII oxidation rate in the light.
“After examining this figure it seems that As(III) is the electron donor and As(V) is the electron acceptor. ” you don’t need the figure to tell you this – basic chemistry determines which can serve as an elecctron donor.
The second half of your comment is spot on though.
Which bacterium? or are you referring to the biofilm. Remember in these inicubations they used biofilm slurries so there may be more than one type of possible metabolim that can be employed depending on the physical (light/dark) and chemical (added electron donor or acceptor) present in the incubation bottle.
In figure 4 the take home message is that AsV can be used as an electron acceptor during chemolithotrophic growth using H2S or H2 as an electron donor/energy source.
The sulfide can also undergo a bunch or abiotic transformations so we probably can add enough to get complete reduction of the AsV to AsIII. It’s also challenging to get good measurements in the sulfide incubations (look at the errors bars for the sulfide bottles).
It would suggest that the organisms are not using acetate as a carbon source in the light. They may be using it as an electron donor in anoxygenic photosynthesis however.
They would have to make a metagenomic libray and screen it with their aoxB PCR product to find a clone containing the full gene. They could then clone the gene into an expression vector and express the gene to figure out what the protein/enzyme it codes for does.
There’s been work since this paper that showed that one or the arrA genes they reported actually functions as an arsenite oxidase – it preforms the reverse reaction of what the sequence match would suggest. They mention this further up the page in paragraph 7.
They’re for two different processes: arsB is used to pump the arsenic out of the cell – costs the cell ATP of PMF to do this; arrA allows the cell to use AsV as an electron acceptor (conserves/releases energy).
Tee chemistry of the water is that is contains a lot of reduced chemical species that can serve as electron donors for chemolithotrophy. These chemolithotrophic oxidations yield more energy of you have a supply of oxygen to use as electron acceptor. The fact that the ponds are shallow means that some oxygen could potentially diffuse into the water and be used as an electron acceptor to fuel oxidation of things like sulfide using oxygen as the electron accepotor.
The authors suggest that a broader diversity of microbes (with a broader range of temperature optima) carry out the reduction but that oxidation is carried out mainly by Ectothiorhopospira strains due to the match in temperature range of PHS-I and the biofilm oxidation.
It could in theory contribute, but they don’t present any data for reduction by PHS-I so I would assume that they tested for it but didn’t detect any activity. They may state this in the text. I don’t remember.
They don’t know “who” is doing the reduction but as long as there isn’t a lot of HGT pr the arrA gene they can use the arrA sequence data to make informed guesses.
The waters are so reducing that there is likely some abiotic reduction of AsV going on but the data show that biological reduction is the main cause of AsV reduction when you supply an electron donor (either sulfide or H2).
H2S is abundant in Mono lake so they wanted to test whether the microbes can use it. H2 can be produced during fermentation which could also occur in the biofilms so they tested that as well.
This is a good question. The simple answer is that the genes or enzymes determine it but the real question is why a pathway that’s pretty well conserved to this point produces this variety of intermediates. I haven’t read much on the topic so I can’t answer the question.
Most likely. There are a few a few bacteria described in the literature that can only get their carbon from hydrocarbons but most hydrocarbonoclastic bacteria can utilize a variety of carbon sources.
Fluoranthene is a PAH. It’s a class 3 carcinogen and therefore a concern. Like many PAHs it can be produced from incomplete combustion or organic matter.
It’s easier to sample terrestrial environments. For anything other than coastal waters you need a boat or ship! All our isolates from fall semester came from soil I sampled at the base of a telephone pole in front of my neighbours house – almost no effort or resources necessary!
There are likely other bacteria that are closely related to C. indicus P73T but I think this paper may be the first C. indicus genome reported. We’ll see in the methods that they compare this genome to the genome or another Celeribacter strain to identify the PAH-related genes.
The details of the method don’t matter. The key details to get from this paragraph are that they are working with 2 different Celeribacter species and that they isolated genomic DNA from them. How they isolated the DNA is irrelevant – you (the class) don’t know enough yet to assess their choice of methods.
I’ll post the reference section so that you can look up the references if you want more details.
Again, google is your friend for these kinds of question but you don’t really need to know what Solexa sequencing is. What you need to take from this paragraph is that (1) they sequenced the entire genome, (2) they identified all the potential protein coding genes, and (3) they identified genomic islands.
Yes. They use the cre-lox system to knock out a gene they think has a role in PAH degradation and then see how the mutant and wild type behave when incubated with PAH.
They don’t know where the fluoranthene genes are located so they chose a genome sequencing-based approach. What other approaches couold they have used? Think about the second paper we read last semester in 314.
Pay attention to the figure legend. It explains the use of the colors. The first 4 rings show the position of diffecent classes of gene in the genome (color key on the right). Rings one and 4 are hard to see because they don’t have a lot of genes but 2 and 3 show the position of genes from various COG (cluster of orthologous groups).
Ring 4 in clack shows the GC skew. Remember that organisms typically have a relatively even GC % across their entire genome. Areas that deviale significantly were probably acquired by horizontal gene transfer from other organisms.
They found a lot of fatty acid synthesis genes, not fatty acids per se. You need multiple genes to synthesize fatty acids. The biochemical pathway have multiple steps depending on what kind of fatty acid you are making. To me 53 genes related to fatty acid metabolism seems reasonable.
The cell won’t dissolve in water! They’re suggesting that P73 produces surfactants that will help it access hydrophobis PAHs etc by solubilizing them in water. They could do BATH or E24 tests like in the first paper we read last semester to confirm this.
This is a good question. We could calculate the energy released by oxidizing fluoranthene completely to CO2 but the yield to Celeribacter would be much less.
They will only knock out the gene they target (0346) but given that many bacterial genes are arranged in operons disrupting one gene will often affect the transcription of genes downstream from it.
Yes. They’re essentially activating the ring. It’s analagous to the ^-carbon phase of glycolysis where you expend 2 ATp to activate glucose before harvesting energy (in the form of ATP and NADH) in the 3-carbon phase.
Yes. There are PCR primers available for some of these dioxygenases so you’ll be able to use a PCR based approach to find some of these genes if we have to wait for the genomes.
We won’t use GC-MS as we don’t have one available in the department but we can look for accumulation of colored metabolites and we can also measure growth on various substrates. We can do single gene transcript analysis of our isolates grown on different substrates.
There are a number of different groups of aromatic dioxygenase (see the tree, fig6?). They’re just saying that previously described fluoranthene dioxygenases were in other branches on the tree.
No, the heavy metals are pollutants as well. They did not use heavy metals. The heavy metals are just as much or more of a problem than the PAHs.
PAHs can be removed when the organisms oxidize them to CO2 when they use them as a carbon source. The metals cannot be removed. They can only be transformed to different oxidation states. We covered this concept in BLY 314 last semester.
That is the assumption. Catabolic genes should not be transcribed (or should be transcribed at an extremely low level) unless the substrate is present.
The cells need a carbon source to grow and make transcripts. They want to compare transcription during growth on PHE to growth on a control/non-PAH substrate and chose glucose.
The BLY 314 class did the isolation and initial characterization of our strains for us last semester. Our job this semester is to find the PAH degradation genes.
Yes, they will. We’ll also send the P. putida RHD that I cloned out for sequencing to make sure it has no mutations before we move it into an expression vector.
They’re using the technique to amplify up the part of the genome that contains the RHD genes so that they can clone, sequence, and study them. They have primers that target part of the RHD but they need to get the full large and small subunit sequences (and maybe some neighbouring genes) so they’re walking out from the area they have primers for.
So at lower concentrations the bacterium van degrade all the PHE but at higher concentrations either the PHE level is toxic or the degradation capacity is maxed out and can’t process all the PHE. We saw somethiing similar in our first paper last semester in 314.
As the genes are transcribed and translated the concentration of substrate (and inducer) goes down due to the activity of the transcribed degradation genes. Less inducer present means less transcription = less mRNA.
Yes, copper increasing transcription of the PHE-degradation genes was unexpected. They suggest that it may be because the cells will get more energy to resist the toxic effects of copper if they increase expression of the phe genes.
Caroline has it. The arrowhead indicated the direction of transcription so we can tell which genes may be co-transcribed (and co-regulated). It’s similar to figure 4 in paper 1.
Yes. The reason they did this comparison is that some plants can be used to bioremediate metals from soil and water. They take up the metals into their tissues and you can permanently remove the metal from the soil by harvesting the plants.
Plants can also enhance microbial activity in soils by (1) increasing soil aeration and (2) releasing fixed carbon into the soil to promote microbial growth.
Yes and no. I’ve added a link to figure 1. It shows three possible initial degradation steps described in the text. Two of them are similar to what we’ve seen before: both molecules of O2 added across a C-C double bond. The third possibility is a mono-oxygenation where only one oxygen molecule is added to the fluorene molecule at carbon #9.
Keep in mind that we’re more interested in the genes than in the details of the biochemical pathway. Focus on the big picture which is that there are three possibilities for the first step in the pathway and that two of them look like what we saw in papers one and two.
Sometimes the initial dioxygenase can attack a wide variety of PAHs but the source bacterium cannot degrade the compound all the way to CO2 because one of the downstream enzymes cannot act on an intermediate.
Yes. The Gram-negatives describes so far have genes that look like they could metabolize fluorene but they can’t grow on it. This could be because they lack a transporter to bring the fluorene int the cell. It could also be a case of sequence similarity misleading us. The true substrate of the described genes may be some other PAH. We should also keep in mind that these initial RHD enzymes often have a relaxed substrate specificity but that the downstream enzymes in the pathway are often more limited in their substrate range.
Also, take a look at the dioxygenase poster outside the door of the lab. You’ll see that Gram-positive and Gram-negative RHDs cluster separately.
They’re working with a gram negative because tat is what grew up when they did an enrichment culture on fluorene. It’s not that they went looking for (or “wanted”) a gram negative, it’s simpy that that is what they found.
See this paper for a discussion of angular versus lateral dioxygenation: https://www.ncbi.nlm.nih.gov/pubmed/12483604. All you really need to know is that they still add two oxygen atoms across a C-C double bond.
The SDS-PAGE allows them to assess whether the cloned proten was made. You usually wouldn’t do it unless you had a problem with the expression experiment (for example no activity against any substrate) and wanted to confirm that the protein was made.
When you put the genes into an expression vector and turn on expression you are over-expressing the protein. The plasmid is a high copy number plasmid. T7 RNA polymerase makes LOADS or mRNA, way more than would be produced normally!
You can’t do the derivitization reaction (which helps with detection on the MS) on a dry substrate. Also, the sample needs to be disolved in liquid to inject int the GC-MS.
They cloned the flnA1A2 genes into a plasmid. They didn’t clone a plasmid. Look at the individual steps described in this paragraph. Break out each step and identify what they did and why?
Yes. They want to test the activity of the enzyme the flnA1A2 genes encode. The way to do that is make an expression clone and put it into E. coli (which lacks RHD genes) and measure the activity of the enzyme towrads various substrates.
The antibiotics ensure that E. coli maintains the plasmid vector. E. coli is lazy and might get rid of the plasmid if we didn’t maintain selection pressure for the plasmid by growing E. coli on media containing the antibiotic.
This is why we refer to antibiotic resistance genes in cloning vectors as selectable markers. They select for cells that contian/maintain the vector.
The big thing to get out of this paragraph is that there was initially a problem with inactive expression clones so they (1) checked that the proteins were made (they were but the MW was a bit off, which they explain), and (2) changed the growth and induction conditions to increase the proportion of FlnA1-FlnA2 that was in the soluble (and likely active) fraction. Step (2) yielded active protein.
That is the most likely explanation. The other possibility is that LB126 cannot bring phenanthrene into the cell. It may have a transporter that brings in fluorene but cannot bring in phenanthrene. Both are large enough that they must pass through transport proteins.
We can’t answer that “why” question based on what is presented here and it’s not important for our understanding of the paper. You’d need an enzyme biochemistry course to get the answer!
It means they got no product from LB126 using those primers. Try to move away from the idea of “worked” vs. “didn’t work.” Focus on what the results tell us which in this case is that the “gram-negative” primers did not yield a product which tells us that the primers were not a good match to the dioxygenase we know that LB126 possesses (we know it has a dioxygenase from previoous work looking at the biochemistry of growth of LB126 on fluorene).
Read the review article I pointed ye to in the previous section. Remember that I told you not to get bogged down in the details of the biochemistry. Focus on the big picture. Which substrates can the flnA1A2 clone oxidize and which can’t it.
It’s probably the only practical way. Given that it is used in agriculture it would be spread over a large area and it would be impractical to remove contaminated soil and groundwater for treatment.
Yes. Unlike us, they don’t have a colored metabolite to help them but by measuring growth in 96-well plates they can identify mutants. It’s not as easy as looking for loss of indigo production but it does work.
Yes. They isolated the gDNA from the mutants, digested it with an enzyme that doesn’t cut the plasposon (which guarantees that they’ll get the mutated genomic region as well) and then ligate, clone and sequence the plasposon integration site.
This paragraph just gives us the growth conditions for all the experiments. You’ll need to read the descriptions of the experiments below to figure out what controls were used for the different experiments.
What do we mean when we say a gene is expressed constitutively? How does constitutive expression compare to inducible expression? How would the graph look for an inducible system?
Not sure I agree with that conclusion based on what is presented in this paragraph. All tey’re doing here is telling us that based on 25 housekeeping genes, KN65.2 falls within the sphingomonads, Novosphingobium in particular.
It probably has more to do with where the pathway was interrupted and how they screend for mutants. They didn’t just look for loss of function, they also examined mutants with reduced growth. Almost all the mutants could break the aromatic ring (released CO2) which suggests that the mutantiosn were for the most part pretty far down the degradation pathway and not in the initial steps.
The language is a bit imprecise but that’s how I read it and it makes sense given that the insertion is in a transporter which presumably brings carbofuran into the cell for degradation.
The question is how would lack of the transporter turn off transcription and translation of the cfdABCDEFGH genes. It makes sense that you don’t make the proteins unless the substrate is present but how do you think that is controlled (if the system is regulated like lac, what is happening at the promoter and operator and what do you think is the effector molecule?)
There is nothing about HGT in this paragraph. They’re just looking at the phylogenetic affiliation (or identity) of KN65.2. They have the genome sequence so they were able to analyse more than just the 16S rRNA gene. They looked at 25 housekeeping proteins which don’t undergo HGT and therefore are useful in determining phylogenetic affiliation.
No purpose. Constitutive expression of a carbofuran catabolic pathway would usually be considered a waste of resources as the species is probably not usually in a carbofuran containing environment.
Figure 2 shows growth on carbofuran. Figure 3 shows either removal (biodegradation) of carbofuran in black circles or mineralisation of carbofuran (where the carbofuran C is oxidized all the way to CO2) in white circles. If the carbofuran is being used as a carbon source we would expect most of it to be oxidised all the way to CO2 (just as glucose that goes through glycolysis and the TCA cycle is). However, the cells might only detoxify carbofuran, where the carbofuran concentration would decrease but we wouldn’t see a stoichiometric formation of CO2. A mutation halfway through the degradation pathway could lead to partial degradation of carbofuran where the black circles go down in value but the white circles don’t increase very much or at all.
They’re all still Novosphingobium. They’re just saying that they had 5 types or classes of mutant (which presumably had mutations at 5 different steps in the carbofuran degradation pathway).
No. I read it as being evidence that it could be a HGT acquired gene – it’s only in one other Novosphingobium isolate but it’s also found in an unrelated bacterium (Brevibacterium).
No. Think about scenarios where a mutation in one gene could affect the expression of other genes. One possibility is when you mutate a regulatory gene but there are others . . .
No, the pink color forms when the only C and N source is carbofuran. It’s a result of using the carbofirnas a N source. When a different N source is included in the media the pink color does not form.
Yes. Because several genes are often transcribed on one mRNA in bacteria a mutation in gene 1 could also knock out genes 2 & 3. Gene 3 might actually initiate the pathway while gene 1 could be for a later step but if you only looked at loss of phenotype you might miss-assign the initial step to the gene 1 product because that is where the plasposon inserted.
All they’re saying in this paragraph is that there is no evidence that the KN65.2 carbofuran degradation genes are plasmid borne. Another bacterium, MS2d, has the genes on a plasmid but that doesn’t appear to be the case for KN65.2.
Yes. It appears to be an indirect effect that is slowing the bug down, probably because of accumulation of a toxic intermediate in methylamine metabolism.
This group of mutants remind us that even when looking at single knockouts we have to consider the organism as a whole.
Yes. The fact that it couldn’t grow on methylamine is interesting given that it had all the methylamine utilization genes. However, this shouldn’t surprise us. It may simply be that it doesn’t have a transporter to bring methylamine into the cell and that the genes are just for metabolism of endogenously produced methylamine.
BLY 314 students should recall the citrate utilization test where some bacteria is citrate negative even through they possesses genes to metabolize citrate. They lack a transporter to bring exogenous citrate into the cell.
This is a good question. They don’t have a specific explanation and just suggest that a toxic metabolite accumulates. It would have to accumulate at a step after CO2 production. Otherwise we wouldn’t see mineralization. It’s not a very satisfactory explanation but would probably require another paper’s worth of research to answer definitively.
No. What they said is that the group III mutants produce only 50% of the CO2 that the wild-type does. Mutations in the cfdH gene reduce CO2 formation by 50%.
What do you mean try a different gene? Do you want to do a directed knockout? How would this help you to understand carbofuran degradation? How would you choose your target gene?
Right now, in this paper, we have a pool of mutants that fall into 5 groups and we’re using the genomic location of the mutations, the phenotypes, and the whole genome sequence to puzzle out as much of the degradation pathway as we can.
I’m not sure what you mean by no effects or which mutants you’re referring to. All the group V mutants accumulate carbofuran phenol. Figure 6 suggests that formation of carbofuran phenol is the first degradation step. In the next step CfdC attacks carbofuran phenol, adding an oxygen to the furan ring, breaking it. If there is a mutation in cfdC, carbofuran should accumulate.
We’d have to do a qRT-PCR to accurately answer this but probably not. Remember that in Figure 1 carbofuran-induced and uninduced cells had the same CO2 production rates.
Yes, when we look at figure 6 (proposed pathway) a number of steps have no enzymes identified, including the initial step(s) that lead to carbofuran phenol formation.
However, the failure to identify that enzyme does not mans that this is a bad paper. It’s an excellent thorough paper that significantly advances our understanding of carbofuran degradation genetics.
Converting the PAHs into TCA cycle intermediates is more efficient that coming up with a completely new metabolic pathway to degrade them. The bacteria just convert then PAh into a compound that can enter a preexisting catabolic pathway.
Good question. You should think about the chemical properties of PAHs vs skin (maybe also the physical properties of the epidermis) surface properties for this question.
Think back to organic chemistry. Are tere differences in stability or reactivity between LMW PAHs (like naphthalene) and HMW PAHs with chrysene or benz [a] pyrene? What distinguishes aromatic hydrocarbons from aliphatic hydrocarbons?
We’ll learn later that the ARHD enzyme has an alpha-3,beta-3 structure. Both subunits are necessary for catalysis. The ARHD also requires the ferredoxin and reductase enzymes for catalysis (they are electron carriers).
We used this approach in lab last week. Our R2A plates contained indole which reveals the presence of an ARHD when it is converted to indigo.
We\’ll use it again this week to help identify our PAH degraders.
One thing to keep in mind as we read this paper is that when we talk about bioremediation of hydrocarbons like PAHs we’re talking about a process in which the PAH is going to be consumed by the bacterium. It will be oxidised all the way to carbon dioxide or partially oxidized and incorporated into cellular biomass. Either way it will be completely removed from the environment.
Pay attention to the media used . When they grew strain CMGCZ they grew it in LB , a rich medium. The initial isolation was done on MSM, a defined minimal medium where Fla was the only carbon and energy source so it took the bacteria a lot longer to form visible colonies.
What does it mean to wash something? Why do you wash something? Here they are washing the bacterial cells before the next step in their experiment. Why?
You’re on the right track. They grow the cells in medium with Fla. This should induce (switch on) expression of the PAH degradation genes. They them transfer the cells to media with ILCO but first they wash the cells to remove any residual Fla. They do this because any Fla that was carried over to the ILCO incubation could cause an underestimate of the ILCO degradation rate.
bead beating is a way to lyse the bacterial cells when you are purifying genomic DNA. We’ll use a combination of heat and chemical lysis when we extract genomic DNA later this semester.
The first enrichment culture contains 10% soil so that they have a large inoculum to increase their chances of isolating a Fla degrader. After that initial enrichment culture has grown on Fla for a while we assume that the FLA degraders have increased in abundance relative to the non-degraders. A larger dilution can be made in the next step because of the initial enrichment step.
One could also use HPLC or an ELISA-based technique but GC is most common for PAH analysis. It is often coupled with mass spectrometry which gives structural information.
They’re talking about two different genes: the 16S rRNA which can be used to identify a bacterium to genus and (sometimes) species and the ARHD gene which encodes part of the first enzyme in the degradation pathway. These two genes have different evolutionary histories so we wouldn’t necessarily expect that they would exhibit the same sequence homologies.
Look up the structures of all three test PAHs. A priori we’d expect Nap to be most easily degraded, why?
In this case, biology (in particular enzyme structure, ability to bind the substrate) trumps chemistry and even though we might consider Nap the “easier” substrate to degrade, the degradation experiment tells us otherwise.
What do you think the yeast extract adds to the medium that may be lacking in the MSM? Probably mostly nitrogen but also other easier to degrade carbon sources.
You can report any % homology. It’s not quite the same as talking about homologous genes and deciding whether they belongf to the same gene family. In this case we know we’re comparing the same gene (16S rRNA) from a number of different species. The usual cut off to consider bacteria members of the same species is that their 16S rRNA gene homology be 97% or greater.
Maybe someone who has taken stats can answer this question? Why perform replicate measurements at all? Why not analyse just one sample for each time point?
The different concentrations let them assess the degradation limits or this Rhodococcus. What’s the highest concentration it can handle, what’s the lowest?
You’re on the right track. PAHs are toxic after all. In theory there should be a concentration at which the bacterium can’t degrade it fast enought o bring the concentration below the toxic level.
In this case the clear zone indicates removal of PAH. The PAHs are not water soluble so one way to add them to growth media is to spray them onto the surface of the agar in an organic solvent that will evaporate off leaving a thin film of PAH behind. This looks a bit cloudy and as it is removed a clear zone forms.
probably because it is found in all microbiology labs! Also because it is an easy way to supplement with all 20 essential amino acids and other growth factors as it is just hydrolysed yeast cells.
See my reply to your comment in the results section. There are a couple of potential factors: first is toxicity, second is that transport systems can become saturated.
Something to keep in mind is that bacteria can engage in horizontal gene transfer. Degradation genes are often carried on plasmids which can move between different species (and genera) of bacteria. Just because two isolates are closely related as revealed by their 16S rRNA gene sequence does not mean that they will have similar activities.
Phylogeny does not determine function for bacteria or archaea.
They used a different primer set to produce the 100-bp product. This means the primers bound to a different part of the Rieske motif than the primers that produce the 78-bp product.
This is true. The background information on PAHs is the same but the focus of the study will differ beyond the fact that they’re studying different genera.
They’re interested in petroleum PAHs because the major sourse of PAHs in marine systems is oil spills, whether from blowouts like the DWH or shipping accidents like with the Exxon Valdez.
I think it’s also cool that a technique we used to identify NP degraders (indigo production) can also be used to identify clones in a genomic library that contain the genes that catalyse the first step in PAH biodegradation (and production of indigo from indole).
Things to look up here. What is an expressin vector? Why colne genes into an expression vector instead of just studying the genes in the clone from the genome library?
Also, pay attention here to the genes they cloned into their expression vector. Why did they clone those particular genes? What proteins/enzymes do they encode?
You should be able to find information on Luria-Bertani agar in he Difco manual posted in the resources section of Sakai. It is often abbreviated LB. I’m not sure what else you need to know other than that it is used to cultivate E. coli but the Difco manual should answer your questions.
I think they give the M9 recipe in a table in chapter 5. It’s an example of a defined medium. Why do you think they would use a defined medium for this experiment?
Think back to BLY 302 and when you learned about Sanger sequencing. How does Sanger sequencing work?
Also, do not ask why particular kits or experimental techniques were used. Ask why the particular experiment was done.
Make sure your comment is attached to the correct paragraph. To answer this question use google! It looks like others have answered the question for you.
Just keep in mind that you don’t have to understand everything about the analytical techniques, you just need to understand why they used them and what information the experiment yielded.
I think we fell down a bit of a rabbit hole here! In any experiment where you are going to do analytical chemistry you want to use the purest chemicals you can because otherwise you might detect a contaminant from your chemicals and erroneously think it was a metabolite produced by your isolate or clone.
What goals did the authors set in the introduction? Are they goals that can be achieved by looking at the whole organism (as in paper 1) or do the require a gene-level study?
In lab we used RNaseA which breaks phosphodiester bonds in RNA between C and U bases. We used it to remove the RNA from our preps.
Restriction enzymes also break phosphodiester bonds but they do it in double stranded DNA and at specific recognition sequences that are 4-10 bp in length.
RNaseA destroys RNA. Restrictionases cleave the genomic DNA into fragments that are a good size for cloning. The LAnder videos discuss this concept well.
The genes will be inserted into the expression plasmids and then E. coli will make the proteins the gens code for and the authors can measure what the proteins do!
The medium is selective if they add an antibiotic to kill all the cells that don’t contain the plasmid. In this case they’re not extracting DNA. They’re going to assay biotransformation of PAHs by E. coli containing cloned genes (on pPhnA) or an empty plasmid. Why did they assay E. coli containing empty plasmid?
So they are not concerned with the yield of the product here. They are concerned simply with the structure of the products. But yes, recovery can be an issue in these experiments especially if you don’t know what the product is (and therefore have to guess at the best way to extract it from the culture supernatant).
Yes, the reason we add the indole to our plates is to detect the activity of the aromatic dioxygenase, which suggests that our bacteria can degrade naphthalene or other PAHs.
The authors state in the last sentence that they contain the same fragment 10.5kb fragent of the A5 genome. This often happens when you make a genome library. The videos in the announcement earlier this week show how this can happen.
They will next subclone different parts of the 10.5kb fragment to get the genome region that contains only the genes they want to study. See how parts B and C of Figure one contain different genes from A. They do this because they want to look at some of the genes individually.
You need to read these papers carefully. The details are very important. It can be helpful to draw diagrams of the various clones they describe rather than just relying on the text.
Part A shows the arrangement of the genes in the genome (and on that 10.5kb Sau3A clone fragment of pH1a).
They sequenced that region and used bioinformatic tools to identify the genes and the restriction enzyme sites.
They used some of the restriction sites to cut out the regions in pH1A that contained the genes they were interested in studying.
For example, digesting pH1a with enzymes P and S yields a fragment that contains the genes encoding the large and small subunites of the ARHD (ph1A1a and phnA2). you can go back ti the methods to see exactly how they constructed plasmids B & C or you can look at the map and figure it out looking at the restriction enzyme sites.
Looking at the annotations for orf 6 & 7 in Table 3 they have nothing to do with PAH metabolism. They may have been \”dropped\” in by a transposase at some time in the past. Remember that bacterial genomes are subject to a lot of \”remodeling\” due to HGT.
They’re not hinting. They’re ascribing function based on DNA sequence identity. This is one of the reasons we sequence DNA, because we can use that information to infer fuinction.
Remember that we see evidence of things like gene duplication all the time in genomes. It could just be that there was duplication of the lsu but not the ssu. Clearly there is a functional ARHD as A5 and the pH1a clone both produce indigo.
We’re just seeing that evolution is not always “tidy!”.
The level of identity can be an indication of how similar the substrate range of the enzymes are. We might predict that PhnA1 will act more like the protein with which it shares 61% aa identity than the ones with which it shares less that 45% identity.
It’s hard to answer “why” questions in biology. Given that the enzyme encoded by A3 interacts with the A1A2 enzymes we’d expect it to bear a similar level of identity as we saw for those components.
You can’t publish a paper about the cool new gene you found without learning a little bit about what it can do!
Basically, you need more than just a BLAST match to assign function. You need to do actual experiments to verify the function the sequence match suggests.
I’m not sure what you’re getting at with this question. Location on the DNA strand? Location on a plasmid vs. a chromosome? Your question needs to be more specific.
Remember that the first step in PAH degradation requires that electrons be supplied to the RHD. The ferredoxin and ferredoxin reductase are the proteins that supply the electrons.
Remember in our intro to metabolisms ferredoxins are a class of electron carrier molecule.
It’s just another way of naming or describing the (A)RHD enzyme. The catalytic sites in the enzyme have iron-sulfur clusters (as do many other enzymes).
The relative location of the genes has more to do with the evolutionary history of the horizontal gene transfer events that introduced the genes to the species and any subsequent genome rearrangements/events that may have occurred.
Habitat won’t directly play a role in where the genes are located. It’s more likely that the genes may have been arranged differently in the ancestor of the Rhodococcus in paper 1 and Cycloclasticus in this paper.
They’re just saying the arrangement and location in A5 are unique compared to what was found in previous studies.
When these PAH degradation genes were first described they were all on plasmids but if you think about what you know about bacterial genomes from BLY 302 there is no reason some of these genes wouldn’t be found in bacterial chromosomes due to transposition events.
The phnA1 gene encodes an RHD-alpha subunit. It doesn’t use any proteins. The branching pattern on the tree tells us which other RHD-alphas it is most closely related to. The fact that it falls outside means that while it’s a true RHDalpha, it differes from the majority of previously described RHDs.
Try not to focus too much on the numbers. We don’t know enough yet to compare 44% identity to say, 60% identity in a meaningful way.
The substrate range of the enzyme is just a characteristic of the enzyme. It doesn’t affect the enzyme itself. It does affect the organism that contains that gene/enzyme, right? It affects what the organism (Cycloclasticus A5 in this case) can use as a substrate for growth and energy production.
Maybe, maybe not. I can’t remember if they told us if A5 can grow on anthracene in the intro or M&M. Growth on anthracene would suggest that another enzyme is present in A5 that initiates degradation of anthracene given that PhnA cannot.
Can anyone describe what is meant by upper and lower pathways? I think this is key to understanding this section. Is the pathway I pasted at the top of the page an upper pathway, a lower pathway or both?
Probably METAtranscriptomics as we’d be looking at the entire community but yes it would be a great tool here especially as the tell us in the next paragraph that they couldn’t amplify true arsenite oxidase genes with their arsenite oxidase PCR primers.
Yes. Some communtiy members are using arsenite as an electron donor during energy generation. Others are using the arsenate generated from arsenite oxudation as an electron acceptor during their metabolism and generating the arsenite. The arsenic is being cycled from AsIII to AsV to AsIII to AsV etc. etc.
It’s the same story that we see with NAD+ and NADH in the cell. NAD+ is reduced to NADH during oxidation of glucose (in glycolysis and TCA cycle) and the NAD+ is regererated when NADH gives up the electrons to the ETC.
Great work seeing the parallel with our pahE PCR “failure” in lab. Sometimes primers that work for some organisms don’t work with the bugs you are studying.
Thank about other strategies they could use to find the arsenite oxidase genes. Sarah Grace had a good suggestionin teh previous paragraph.
So they are going to do two types of analysis on the biofilms.
1) DNA analysis – to see “who” is there and “what” they can do.
2) Incubations – add the biofilm to media and measure the various transformations (of AsIII, AsV, various carbon and electron sources) the biofilms carry out.
They describe a number of different experiments here. Some look at As transformations. Others look at C-use. It will make more sense when you see the data in the next section.
In the first experiment they are looking at arsenic transformations. The key thing to know here is that radioactive As is added in very small amounts. It is a tracer. Most of the As in the experiment is unlabeled.
Remember that in the introduction they told us that these biofilms show evidence of respiratory As(V) reduction, photosynthetic anaerobic As(III) oxidation, and aerobic As(III) oxidation. This section describes the incubation experiments that lead to those insights.
Journals require that authors group similar experiments in the same section. Part of your job as a reader is to separate them out again. Take notes as you read this paragraph. Pay attentions to electron donor and electron acceptor as well as carbon and energy source.
The physical and chemical description of the site gives us the information we need to form hypotheses about what kind of metabolisms might be found there.
Hang has it. They’re preserving the samples in an inactive state so that they can do their experiments later with essentially unaltered samples that represent the state the biofilms were in when they originally sampled.
We don’t care about evolution here. We want to determine what the bacteria/archaea can do with the various C, E, and e- sources – how they use them in their metabolism.
They pool three independent replicates in case the PCRs behaved differently. It should give them a more representative sample of the community in the biofilm than if they just did one PCR.
With only 96 total clones analysed they discarded the singlets because they’re not confident how representative they are. They just focused on the more abundant groups. It’s not critical for our understanding of the paper.
Think about what they will do with the samples after storage. For incubations they store at 4-5C (slows things doown but doesn’t kill the cells. For DNA analyses they store at -80 – kills the cells but preserves the DNA.
Remember that when we discussed genome sequencing we talked about how a certain % of the orfs identified in a genome either have no match in GenBank or the sequence they match has no annotated function.
With degenerate PCR primers you often amplify non-target sequences and many of these have no match in GenBank.
They include killed controls in order to confirm that any transformations they see are a result of microbial activity and not spontaneous chemical reactions.
The amount of light is constant. In the light the AsII goes down and AsV gos up, so AsIII is being oxidized to AsV. What possible process(es) could be using the electrons removed from the AsIII. Make a list!
As you look at this graph think about what process(es) could take AsV and reduce it to AsIII in the dark. AsV -> AsIII is a reduction. The AsV is being used as an electron acceptor by what possible processes?
Lastly keep inmind that these incubations were carried out in sealed bottles (i.e. a batch culture). Over time waste products accumulate in sealed systems and can “slow” down microbial metabolism.
Remember that it’s not just one organism. it’s a community. They can coexist by using different electron donors and acceptors in their metabolism and by supplying engaging in syntrophic interactions where one organism’s waste is another’s fuel.
Addressing both Erica and Christy’s question. One narrow optimum temperature for oxidation might suggest that it’s only one organism that is carrying out this process. The broad temperature range for reduction could be because a variety of different bugs are reducing AsV (each with a different temperature optimum).
The PCR and sequencing of the reduction and oxidation genes could help answer this. if there is just one reduction gene sequence it would suggest just one bug does the reduction. If there are a variety of reduction genes, many bugs reduce AsV.
It’s “good” in that it’s always nice when a follow-up study confirms the findings of a previous study.
Also, as we discussed in class, isolates are not always ecologically relevant. In this case the culture independent 16S rRNA analysis confirms that their previous isolate is in fact a major player in these biofilms.
What do you mean by very similar? They\’re mirror images of one another. In one AsV decreases and in the other AsIII increases.
Why does this happen? The AsV is reduced to AsIII, so AsV goes down and AsIII goes up.
The critical thin to identify here is where the electrons used to reduce AsV to AsIII come from. What else decreases in the top panel of the graph?
Not quite sure what you’re getting at here. Remember that 16S genes are not usually subjected to HGT. The finding of only one type of bacterium in the biofilm simply suggests that the biofilm is essentially a monoculture on the bacterial side.
HGT might play a role in arsenate reduction but may not. remember that this ability evolved early in life in earth and would be expected to be found in many phyla (at least those still living in places like Mono lake).
Mono lake experiences temperature extremes. In summer the temperature can range from 6 to 36C. It’s high in the Sierra’s and gets >60 inches of snow a year.
Until they do the experiment all they have is a hypothesis. They had to do the experiment to gather evidence to support or reject the hypothesis that H2 and or Sulfide oxidation was a major component of the lake biology.
But there could be oxidation of acetate in combination with reduction of a different electron acceptor… The key is the second sentence of the next paragraph…
They’re saying that acetate is not oxidized in conjunction with AsV reduction. In the next paragraph they tell us that acetate is oxidized under a different metabolic scenario.
They subsequently found (after doing some genomics) that the arsenite oxidase in this system \”looks\” like an arsenate reductase sequence-wise. It has some key substitutions that make it work in reverse!
Equal and opposite reduction of what? They saw acetate oxidation in the light but not in the dark. Where do the electrons from acetate end up in the light?
So in photoheterotrphy the energy comes from light (photo) but the C comes from organic molecules (hetero). A bit confusing because we often think of heterotrophs meeting their C, e-, and E needs from an organic molecule bur microbes are very metabolically versatile.
Hopefully this drives home that it’s not just photosynthetic organisms that fix CO2 into biomolecules. Chemolithoautotrophs do it as well. Some use the Calvin cycle but others use some of the other carbon fixation pathways we touched upon in lecture.
Sometimes bioremediation occurs without any manipulation of environmental conditions. for example in the aftermath of the DWH blowout in 2010 microbes in the GoM degraded a lot of the released oil without any special measures being taken.
Rates of bioremediation are quite variable and depend on the exact substance being degraded, its concentration and other environmental conditions (think about things like temperature and availability o essential nutrients such as n, P, K and Fe which could limit microbial growth).
No control need for collecting the sediment as this is a discovery type study. They’ll run controls later though when they’re measuring PAH degradation etc.
They’re doing an enrichment culture, like Beijerink in chapter 1 and like we’ll do in lab next week. Read carefully – they add the PAH as a sole source of carbon so only PAH metabolisers will be able to grow and reproduce.
They’re not comparing the isolates to E. coli. The numbers in the primer names (27 and 1492) refer to the position of the primers in the sequence of the E. coli 16S rRNA gene.
What does pH measure? What does conductivity measure? Why would these affet living organisms? Think about the basics you learned in oter classes about proteins and enzymes.
There’s wave action over the top layers of sediments in the ocean. Also, various animals (worms etc0 disturb the sediment allowing oxygen exchange into the sediments. by shaking at 100rom they mimic this. They need this gas exchange because they’re looking at aerobic PAH degradation.
One reason is the size of the gene (~1500bp). It’s easily amplified by PCR and gives sufficient information for good phylogenetic discrimination. The 5S rRNA is only ~120bp and the 23S is ~3000 bp.
It allows them to graphically depict the relationship of their isolates to the entire diversity of the bacteria. It can be more valuable than simply looking at the information the BLAST search gives you (% identity to closest relative). For example, it’s easier to see on a tree if all yor isolates are closely related to one another.
Read my introduction to this section. Focus on the big picture – yo don’t need to understand all the little details, just why they did each experiments. Most comments on this section got way to bogged down in technical details.
Take a look at the map (figure 1 in the previous section). It may help explain the physico-chemical differences between sites. Keep in mind that conductivity is related to salinity. The key thing in this paragraph is that they sampled 3 different sites – all the sites had detectable PAH (so they would reasonably expect to find PAH degraders) but the sites had different nutrient profiles which could limit bacterial productivity (PAH can supply C and energy but they need N, P, K etc. to build proteins, nucleic acids etc.).
This is a good observation. An isolate with a wide substrate range would be more “useful” than one with a narrow substrate range for bioremediation as crude oil is a complex mixture of hydrocarbons.
Keep in mind that in nature we have a community of microbes with different abilities carrying out bioremediation. We don’t rely on just one “superbug” to do all the bioremediation work where PAHs and crude oil are concerned.
Most often we rely on bacteria that are already present and either add nutrients (N, P, K etc) that may be limiting their metabolism or add a supplemental carbon source that allows them to grow faster and co-metabolise the PAH (but not rely on only the PAH for growth which would be slower).
That could be true. it could also be that the media they used in lab is not the best choice for growing these bacteria – we discussed this in lecture when we talked about potential explanations for the great plate count anomaly.
Gram status shouldn’t make a difference. Gram negatives are often faster growing (probably just an artifact of the media we use in labs) but it doesn’t mean that they will degrade PAHs faster. There is some evidence to suggest that Gram positives like Mycobacteria are better at degrading HMW PAHs.
We use cocci and bacilli to describe cell morphology, not colony morphology. For panels 1-2 of figure 2 you’d use the terminology in the lab 1 protocol and describe colony shape, margin, elevation etc.
Based on 16S rRNA gene homologywe’d classify them as the same secies (rule of thumb is >97% identity is the same species). However, we need to keep in mind that they didn’t sequence the entire 16S gene, just part of it.
Also, the 16s gene is just one marker. It’s not perfet and 100% 16S rRNA sequence identity doesn’t mean that the two strains are identical throughout the genome, especially as bacteria can engage in horizontal gene transfer via conjugation/ transduction/ transformation and “pick up” genes from their environment.
Doing a partial sequence was probably partly a budget issue. Science costs money! Sometimes you don’t have enough to do a perfect experiment but you can still generate good, publication quality data.
We’ll do partial 16s sequences for our isolates this semester. They’ll allow us to identify our bacteria to at least genus level and probably species level.
Do you know what the KEGG model is? It’s the Kyoto encyclopedia of genes and genomes. It’s useful for genomic and biochemical studies. Why do you think they’re using it here?
This is a good question. Bacteria are more complicated to analyse that plants or animals as function does not always follow phylogeny (because of HGT). However, identity and relatedness is still a good place to start.
So, for Ochrobactrum there’s some previous work showing this geneus can degrade PAHs. Their work asdds to our understanding of the role of Ochrobactrum in degrading PAHs.
The Cupriavidus result may be the more interesting as they tell us not a lot has been published on PAH degradation by this strain. This art of the paper represents “new” knowledge.
A clear example of how a picture can be worth 1000 words. Note how for LC there is no difference in branch length (horizontal axis0 between it and the other species on that branch – they’re identical.
However, isolate LA is on a much longer branch from it’s closest relative, indicating it’s not as closely related.
It’s been isolated from a lot of cystic fibrosis lungs. Genomic studies have shown that some Pa isolates have a lot of pathogenicity factors. However, we’d want a lot more data that just a 16S sequence to conclude that their LC isolate is a human pathogen.
There’s a small rise at the end of the graph, yes, but we needn’t be concerned with it. We’re just interested in the 15 minute peak for (a0 as that is where phenanthrene elutes and the 28 minute peak for (b) as that is where pyrene elutes. Everything else is irrelevant.
I’d encourage all f you to draw the structures of these compounds and revisit your organic chemistry texts and think about the properties of aromatic compounds.
A report is a previous publication an there is no previous publication reporting that Pc degrades phenanthrene. there are previous reports that it degrades other substances but not phenanthrene. this is the first report of it degrading phenanthrene.
PAHs are not totally water soluble. They form tiny particles in an aqueous solution like growth media. this means they increase OD in the beginning (before the bacteria start to degrade them) but as they are degraded they contribute less to OD so the PAH contribution to OD decreases as time goes on.
Naphthalene is the simplest PAH – only two rings. The larger PAHs with more rings tend to be much more toxic and more difficult to degrade. However, the initial step in aerobic degradation of all PAHs, addition of both molecules of molecular oxygen across a C-C double bond, is the same.
Think about differences between deep-sea and shallow water environments. Would you expect coastal (shallow water) bacteria to thrive in the deep? Why or why not? Temperature is a factor but there\’s another big difference that you\’re missing.
You could click on the Pearson et al link and see what that paper has to say about the topic. It migth help or it might be too technical for us in BLY 314. At most I\’d read the abstract and introduction.
Read the first sentence of the paragraph really carefully. It is the key! The technique is also described in our textbook. we won’t cover it for a while but you can look ahead.
Is the overall goal of the study to degrade PAHs? I don’t think so. They want to study the diversity of the hydrocarbon-degrading bacteria at Guaymas. How will SIP help them do that?
Sort off, in that the hydrocarbons originate in the deep subsurface and are moving up into the deep waters of the ocean. However, the physical conditions, i.e. temperature, are quite different which probably has an effect on both hydrocarbon composition and microbial community composition.
Temperature can act as a selective factor – favouring growth of some organisms and inhibiting others. The vent fluids may also affect what grows where given that they have a different chemical composition than “normal” seawater.
They’re basically doing a survey so they grab samples from a variety of different locations to see what’s there. A control sample would be at some distance from the vent where there was no vent effect.
Adding the PAH makes the media cloudy. If the bacteria metabolize (and remove) the PAH the media turn clear. We’ll do something similar in lab 5 when we look at metabolism of starch and of milk proteins.
So using isotopes (both stable and radio isotopes) allow us to tell if an organisms uses a substrate labeled with that isotope.
In both cases the labeled carbon has two possible fates – it could be partially oxidised and incroporated into cellular biomass or in could be completely oxidised to CO2.
So the key here is that they want to separate the DNA from cells that incorporated the 13C-labeled PAH from the DNA of the cells that did not incorporate the labeled PAH. The labeled DNA will be heavier because it contains C-13 instead of C-12.
The E.coli DNA is unlabeled and serves as a standard for unlabeled DNA. It should migrate to the same spot in the tube as the DNA from the unlabeled sediment bacteria.
They can use 16S PCR and sequencing to verify that they properly separated the C-12 and C-13 DNA because there should be no E. coli in the C-13 DNA because E. coli doesn’t live in vent sediments.
Presumably they got PHE degradation in their earlier incubations and decided to try to isolate PAH degraders from PHE enrichment cultures. Also, the fact that it was one of the PAHs they detected in situ made it a good choice.
People often use naphthalene for these experiments so may they wanted something a little different to see if they would get different bacteria.
They first grow them in pyruvate to get a lot of biomass. They wash the cells to remove any residual pyruvate so that when they add the 14C-PHE the cells are “forecd” to use it because all the pyruvate would be removed.
Microbes will always want to use the easy substrate (gulcose before lactose and pyruvate before phenanthrene or any other PAH).
Sequencing of ribosomal RNA is a common tool to identify/categorise organisms (16S for bacteria and archaea) and 18S for eukaryotes. Organelle genes (for example cytochrome oxidase) are also used in studies of eukaryotes.
For bacteria, 16S rRNA sequences that share 97% identity or higher are considered the same species. We’ll spend teh next 3 weeks in lab working on generating 16S rRNA sequences for our NP isolates.
You can find out by seaching the genera in PubMed or another scientific database. As far as I know Cycloclasticus has only been reported from marine environments. I think Halomonas has also been found in saline terrestrial environments as well as marine environments.
Draw it out. I’ve done it for you here but you should all get in the habit of doing this if the text is confusing you. I imagine Madelyn is not the only one confused by this section!
The bacteria don’t use the sulfide instead of oxygen. They use sulfate (oxidised) as an electron acceptor instead of oxygen and produce sulfide.
The last sentence tells us why they focused on PHE in their experiments – because it was the PAH that was mineralised (oxidised to CO2) to the greatest extent by the 4567-24 sediment sample.
Yes, this graph makes clear that they should focus on the 4567-24 sample (open bars) and use PHE as their model PAH because it shows the greatest mineralisation.
Yes. They used the C-12 and C-14 incubations to track the degradation or the C13 PAH. They want to isolate the C-13 labeled DNA from the bacteria that were the first users of teh C13-PAH. They don’t want the label distributed across multiple trophic levels.
They also don’t want to “waste” their C-13 labeled sample so they use the other 2 incubations to track PAH uptake and mineralisation.
Keep in mind that they are comparing killed controls (where they expect no PAH removal/CO2 generation) with live experiments (where they expect both PAH removal and CO2 generation). The circles measure one thing the squares another. A good exercise is to predict what the graph should look like before you look at it.
For the circles they’re measuring the accumulation of CO2 from mineralisation (oxidation/degradation) or the PAH. In the live incubations this line should go up over time and stay flat (and near zero b/c there are no bugs to degrade the PAH) for the killed control.
For the squares they’re measuring the PAH directly. For the live incubations this line should go down over time if degradation occurs. The killed control should be flat but high (ie the amount of PAH added initially).
So they centrifuge the DNA in order to separate the heavy C13 DNA (from the phenanthrene degraders) from the light C12 DNA of teh non-degraders (and their E. coli control).
Each lane in the gel represents a different layer (fraction) from the CsCl gradient. Notice that the fraction son the left contain different bands than the fractions on the right and that the middle fractions are a mixture. This (and the presence of the E. coli band in only the right-hand fractions tell us that they did a good job separating the C13 DNA from the C12 DNA.
They’re saying their culture work supported the SIP work. They found Cycloclasticus 16S sequences with SIP and they were able to isolate Cycloclasticus from their enrichment cultures.
I think this is also an encouraging result in that naturally occurring bacteria exist that can naturally remediate these pollutants. Also, the system is resilient – many differnt genera seem to be able to degrade PAHs. We don’t need to engineer any superbugs!
As Abrianna reported, neighbour joining is very fast so it’s very popular. We’ll use it and a few other algorithms in lab once we get teh 16s rRNA sequences of our isolates.
I read this as they can only gow on PAHs if they’re obligate degraders whereas the non-obligate degraders can grow on a wide variety of carbon sources.
Yes, they didn’t directly show PAH degradation by Cycloclasticus. The showed PAH carbon in Cycloclasticus DNA using SIP but another organism could have metabolised the PAH and Cycloclasticus could have consumed that organism. This is why they spent a lot of time figuring when to isolate the DNA from the SIP experiment.
The definitive experiment would be to incubate their Cycloclasticus isolate with PAH as teh sole C and E source and measure PAH removal, mineralisation and incorporation into Cycloclasticus.
Remember in BLY 302 you learned that bacteria can acquire genes from many sources. They undergo horizontal gene transfer (from other bacteria, viruses, etc) as well as vertical (from parent) so bacteria can be very closely related as determined by 16S rRNA phylogenies but have different phenotypes (due to the genes they pick up via horizontal gene transfer.
A good question. We’re often most interested in the toxic effects of substances and overlook (at least initially) that there can also be non-toxic uses of them (or useful applications of their toxicity in things like pest control).
Yes and elemental selenium is a solid so it’s not as bioavailable as the water soluble selenate and selenite. We can think of this system as a win-win – the bacteria can survive without O2 by using, for example, selenate, as an alternate electron acceptor and detoxifying the environment at the same time.
The earth is an oxidizing environment (exposed iron rusts etc) so it could be that abiotic processes re-oxidize the reduced selenium. It could also be that there are bacteria out there that can use reduced selenium species as electron donors (get their electrons and energy from oxidation of reduced forms of selenium instead of from preformed organic molecules like glucose).
Yes but with one key difference. In bioremediation of organic molecules like PAHs they can be completely removed from the environment by oxidation all the way to CO2. In biomineralization the element remains in the environment but it is immobilized and much less bioavailable because it is now and insoluble solid mineral.
Look at the reaction they’re looking for. You should be able to tell whether it is more likely to occur with or without oxygen based on how the cell is using the selenium.
Excellent observation. They definitely won’t be able to use K-12 as a host for their clones or as a negative control but there may be other strains of E. coli than don’t reduce selenate that they’ll be able to use.
This is a general methods paragraph that describes the various media they used. We’ll find details of when the media were used in the subsequent sections. Sometimes they’ll tell us explicitly, other times we’ll have to figure it out from this paragraph and the information in table 1.
They wanted to screen the library for clones that contained DNA that conferred the ability to reduce selenate onto the E. coli host. Therefore, they first screened various E. coli hosts for the ability to reduce selenate. They needed a selenate reductiion-negative host.
Review section 12.4 in the textbook (cassette mutagenesis) for help with this section. You don’t really need to understand the details, just that they are trying to knock the fnr gene out of the wild type E. cloacae.
This is not what I want you to focus on. You are getting bogged down in the technical details! Focus on the big picture. We don’t care how they isolated the DNA we care why they isolated it – what did they do with it and why did they do it?
See figure 12.8 in the text or time travel back to the pGLO lab in BLY 121 if you took the 121 lab here. What would happen if you didn’t include the antibiotic that selects for the cosmid in the agar?
It may help to review the section on cassette mutagenesis in the text and to cartoon out the experiment. The knockouts will only carry one of the antibiotic resistance genes from the sacB based vector (The sacB-based vector suicides when you add sucrose to the growth media).
Their goal is to find genes involved in selenate reduction. Based on their knocking out fnr and then complementing fnr it seems that fnr has something to do with selenate reduction by E. cloacae.
Their clone produces a product so they analyse the product.
Remember there is no degradation of Se, there is only transformation from one form to another. The Se is never removed/degraded the way an organic compound can be removed by oxidation to CO2.
Cosmids can accept larger DNA fragments than plasmids so when you make a genomic library you don’t need as many clones as each clone contains a larger piece of DNA.
You need to pay attention to the names here. E. coli S17-1 does NOT reduce selenate. They told us this in the methods. The lack of selenate reduction is why they were able to use it as a host to screen clones for genes that confer the ability to reduce selenate.
Also, S17-1 plus the empty cosmid vector (pPLAFR3) does NOT reduce selenate.
However, the red colony/clone that contained the cosmid pECL1e does reduce selenate. The conclusion is that one of the genes inserted into the pLAFR3 cosmid to yield the pECL1e cosmid confers the ability to reduce selenate on the E. coli S17-1 host.
No, the graph shows that the pLAFR3 clone does not reduce selenate at all. The error bars overlap for the open boxes meaning there is no change in selenate concentration.
In contrast pECL1e reduces selenate at a ~constant rate for the first ~35 hours and then plateaus.
Go back to the introduction and see what the authors told us about the properties of selenium and the effects different forms of selenium can have on living cells. Some organisms can use selenate as a terminal electron acceptor for growth. Are there other reasons an organism might reduce selenate to selenium? Hint: think about bioavailability.
Fnr is a regulatory protein. It turns on the E. coli selenate reductase.
E. coli has the enzyme but some strains like S17-1 lack a functional fnr gene to turn it on while other strains like K-12 have a functional regulator and reduce selenate without the need for the addition of an fnr containing plasmid.
Yes! The native bacteria can carry out bioremediation (by using the diesel as a carbon and energy source) in Antarctica and they could also be useful in other locations.
Bacteria in natural environments are often nutrient limited (often N or Fe-limited). They have plenty of carbon from the oil/diesel but they lack some of the essential elements and can’t make the enzymes they need to degrade the oil. One way to speed up bioremediation in-situ is to add nutrients –essentially add fertilizer like you would in your garden.
It may have to do with the length of their field season or expedition. Many scientific expeditions to Antarctica are 3+ months long. You collect samples while you’re there and do most of the analysis when you return to your university.
In general, even bacteria isolated in cold environments can grow well (and often faster) at higher temperatures. hopefully, they’ll tell us in the next section what temperature was best for their isolates.
It depends on what you want to know. Sanger is preferred for sequencing a single gene (for example to sequence the 16S to identify a bacterial isolate). NGS is preferred for sequencing an entire genome but would be overkill for a single gene. We’ll talk about this more in class when we get to Genomics in chapter 7.
They’re talking about sequencing the genome here. One of the tools they used to analyse the sequence data was the KEGG database (Kyoto Encyclopedia of Genes and Genomes) which allows you to map biochemical pathways onto your isolate’s genome by sequence holology. The paper describing KEGG was published in 2000 (in the early days of genomics) but it’s been updated continuously since then.
Their goal was to isolate PAH degraders so 4 contaminated sites was enough to give them a strong probability of isolating at least one degrader (we probably got >100 blue colonies from my midtown Mobile telephone pole soil!) Previous research suggests that even one contaminated sample will yield multiple PAH degrading bacteria.
They answer your question in the last sentence of the paragraph – they pick those that metabolized the greatest amount of phenanthrene (i.e. the best phenanthrene degraders).
Most likely they grew them in flasks in a shaker incubator, probably at ~200rpm. This is sufficient to keep the cultures aerated. When I grew the cultures used in lab 3 I grew them at 200rpm and 35C. Without shaking the cultures would quickly become oxygen depleted and not grow as well.
Testing for biofilm formation is just another phenotypic test that gives them some more information about the bacteria. It may or may not make them better PAH degraders.
They picked colonies off a bunch of plates that they incubated at 3 different temperatures. They probably had as many or more spread plates than our entire BLY 314 lab generated in lab 2!
They measured phenanthrene degradation by looking for a decrease in “phenanthrene fluorescence” which is fluorescence at a particular wavelength that they attribute to phenanthrene but could also come from other compounds. If the the fluorescence in their phenanthrene degradation incubations (1) stays the same or (2) increases they exclude those isolates because they (1) didn’t degrade the phenanthrene or (2) produced some other compound that fluoresces at the same wavelength as phenanthrene.
Focus on interpreting the actual data presented for now. How the bacteria acquired the ability to metabolize phenanthrene is no the focus of this figure (although it is an interesting question and on that the genomic data later on in the paper will address).
Adherence to the carbon and energy source seems like a good strategy for enhancing degradation of the carbon source. However, there are others. For example, bacteria can produce surfactants to increase the bioavailability of a PAH. They can release exoenzymes that degrade the carbon source externally to the cell and take up the metabolites. Remember the siderophores we discussed in lecture. The bacteria release them to scavenge for iron and then take them back into the cell.
It would be nice if they used the same units for their in-lab experiments and the soil Cd concentrations from various environments. Maybe someone will do the computation for us. You can assume that 1mL = 1g.
The beauty of tis study is that it shows bacteria capable of bioremediation are already there. They may however be nutrient limited in terms of their N, P, K, Fe etc. needs. It’s usually easier to get approval for adding nutrients (fertilizer) than for adding organisms.
During the aftermath of the Exxon Valdez oil spill one of the approached they took was fertilizing the oiled beaches. It was much more effective (and much safer for the environment) than power-washing the beaches.
We learn oil floats on water, right. But if enough “stuff” sticks to it it can sink once it is denser than seawater.
There’s also naturally occurring oil at the seafloor in “seep” environments where oil from below leaks out onto the seafloor. The “natural” oil spills may be a source of the oil-degrading bacteria that respond to oil spills.
Yes. When organisms consume a food (C) source some of the food is respired all the way to CO2 but some is diverted into biomolecules to build new cells.
Average temperature at the sea floor is 4C which is probably why they chose 4C as one of their incubation temperatures. This was also the temperature of the sediment in core 4567-24.
Based on this, any thought on why they picked 21C as their second temperature?
The oil is filtering up from below the seafloor, not down from the sea surface. remember from the introduction that the Guaymas basin contains a lot of hydrocarbon seeps where oil percolates up to the seafloor from below.
Average temperature at the sea floor is 4C which is probably why they chose 4C as one of their incubation temperatures. This was also the temperature of the sediment in core 4567-24.
Based on this, any thought on why they picked 21C as their second temperature?
Look at what they put in the flask. They put in C-14-Phe, a radio isotope, so they measured the radioactivity by liquid scintillation counting.
They ran parallel incubations, one with C-13 (for SIP) and one with C-14 to measure metabolism of the phe. Why? because they didn’t want to sacrifice the C-13 flask just to measure utilization of Phe. They wanted to save the all the C-13 flasks for DNA extraction.
We don’t know if they’re psychrophiles or psychrotolerant form what they’ve described so far. They’d have to test the growth rate over a wide variety of temperatures before they could determine that the bacteria are psychrophilic.
The key is not that C-14 is heavier, it’s that it is radioactive and easier to measure (and also a C-14 labeled compound is often cheaper to use in an experiment than a C-13 labeled compound as you can use way less of the radiolabeled compound as you can detect it at way lower concentrations than the C-13 compound).
You need to read this section really carefully to track what they are using the different forms of Phenanthrene for. Try drawing out a flow chart.
Basically the C-13 flasks will be used for SIP. The C-12 and C14 flasks are being used to figure out how much phenanthrene is being metabolised and if it is being metabolised to CO2 or into biomass which allows them to determine the best time to collect the C-13 labeled cells.
I’ve posted a few short videos explaining the experimental set-up in the Paper discussion module on Canvas. This diagram may also help.
Yes, the E. coli DNA will get mixed up with the unlabeled DNA but it doesn’t matter. They only want the DNA from the labeled cells (which metabolised the labeled phenanthrene). Adding the unlabeled E. coli DNA will let them determine whether they did a good job separating the labeled and unlabeled DNA.
I’m not sure about your numbers but basically, yes. The DNA sample (mixture of heavy and light) is put on top of the CsCl solution, and then the tube is spun at a very high speed . The CsCl molecules become densely packed toward the bottom, so even layers of different densities form. The DNA moves to the CsCl layer that matches its density allowing the heavy and light DNA to separate. It looks something like the picture here.
I don’t think anything went wrong pre se. The bacteria in their samples possessed enzymes that allowed them to degrade naphthalene and phenanthrene but not the other PAHs. A general rule of thumb is that smaller PAHs are easier to degrade than larger PAHs. Water solubility (and therefore availability to the cell) is also important.
Not exactly. Your overall conclusion is correct but your pth to the conclusion is not quite right!
The DGGE gel is separating the 16S products (which are all the same size) based on their GC content. Each lane in the gel corresponds to a different layer (fraction) of the CsCl gradient used to separate the heavy DNA from the light DNA.
Look at the line graph. It shows 2 peaks, the first for the heavy DNA and the second for the light DNA.
What does the presence or absence of the E. coli band in the gel image tell us?
This was surprising but the difference fact that the oil exposed cores were essentially anoxic (sulfide adapted) may explain this. As Shelby said earlier, doing the incubations under anaerobic conditions might have yielded better degradation for the 4571-2 core sediments.
11 days would be really long for our bacteria i the lab as we usually provide them a lot of carbon but for deep sea bacteria 11 days is not very long. The key here is that they chose 11 days based on the data in the graph.
Remember this is a paper about stable isotope probing which is using a stable isotope, C-13, to identify the bacteria using the 13C-Phenanthrene.
“Normal” carbon has a molecular weight of 12, Bacteria that grow on the 13C phenanthrene will have 13C DNA which is heavier than 12C-DNA. They can use centrifugation to separate the heavy and light DNA to identify the phenanthrene using bacteria from the
You were already exposed to this type of experiment in Genetics when you learned about Meselson and Stahl’s experiments showing the semiconservative nature of DNA replication.
Yes, they used the acid to kill the bacteria in the sediment.
In the last paper they used E. coli as a control in their incubations with pure cultures. The only difference was the use of E. coli instead of one of their isolates.
Here, they “kill” the sediment so that the only difference is whether the cells are dead or alive. They wanted to retain the sediment in case there was something in the sediment that might react with the phenanthrene (althoug one might argur that adding the acid may have changed the chemical properties of the sediment).
Check out my comments and videos on the experimental set-up. Basically the stable and radio labelled isotopes are expensive so they do some experiments with unlabeled PAH.
Mostly correct! They only analyse 16S rRNA sequences in this paper. One 16s sequence dominated the sequence data. That sequence matched a sequence from a previously studies Cycloclasticus species which degrades PAHs.
They share a common ancestor in the sense that they’re all proteobacteria but they’re not very closely related. If we sequenced the genomes we’d probably see dramatically different genome content (although due to horizontal gene transfer they might also have similar PAH degradation genes).
Jordan is correct. you can figure it out by looking at the redox couple information presented in the paragraph. remember these couples are always written oxidized/reduced substance so AsV is the more oxidized substance and therefore functions as an electron acceptor, while AsIII is the more reduced substance and ca serve as an electron donor.
They’re using nitrification/denitrification as an example or model for what happens with arsenic in Mono Lake. They are not talking about nitrification/denitrifincation in Mono Lake.
Yes. The microbes use something other than water as e- donor. Given that they just told us about the arsenate/arsenite redox couple maybe the anoxygenic phototrophs can use the reduced member of the couple as an electron donor.
A good question. Why do you think our pahE PCR failed in lab? We’re pretty sure our bacteria are PAH degraders (based on indigo production) so they should have pahE genes.
to a degree, yes. Different photosynthetic bacteria make different accessory pigments so we can use color as a proxy for the various groups. For example, in oceanography we often use pigment analysis to determine which photosynthetic groups are present.
They probably have monthly sampling trips to Mono lake as it is a well studied system. Also, one might find different microbial groups dominating at different times of year. It’s not really important for this study but there are a lot of other papers studied on this system.
The pH of their salt solution must have been above 9.3 before they added the HCl.
Keep it simple. Remember to focus on the big picture, not the minutiae. How they made the media isn’t really important to the big picture. The fact that they made a media that resembled the composition of the Mono Lake waters is the key detail here.
Read more closely. They found matches for the 16S rRNA sequences. Some of the arrA and all of the aoxB clones did not yield matches. What does the latter finding tell you?
Think about how PCR works. You design primers to previously sequenced genes and use them to look for the same gene in other organisms. The arrA and aoxB primers are a concensus and not usually perfect matches. Would you expect the primers to amplify the gene from all other bacteria? probably not.
Protein coding genes are not usually as well conserved as the 16S rRNA gene so they can be more difficult to amplify.
Keep in mind that the bottles contain a finite amount of As that is cycling between AsV and AsIII. Also, if you look at the graph they added the As as AsIII in the beginning.
Your job is to note the changes in AsIII and AsV concentrations and figure out what biological (or abiotic) processes could be causing these changes.
Keep in mind that they’re doing this experiment in sealed bottles where substrates can run out and toxins can accumulate. It’s not a perfect simulation of the natural conditions which are more like an open container where new substrates can enter and waste can diffuse away.
Do you think the reaction is slower in the light or are we looking at a net rate? (Could oxidation and reduction occur at the same time? and is there a reason we would see the two reactions occurring in the light but not in the dark? )
Basically, they’re saying that the sulfide and arsenic are participating in many different chemical reactions and that because of this there are a variety of “As-S” intermediates present at any time which leads to errors/variability in their measurements.
Think about the genomic and molecular biology techniques we learned about. Could we apply any of them to this system? Metagenomics could work as the community is not very diverse and it should be possible to get near complete genome sequences for the dominant members. Arsenite oxidase genes that did not ampplify with the primers they used might be detected by sequence homology in metagenomic sequences.
Also, arsenite oxidation was carried out by essentially a pure culture (Ectothiorhodospira-like). They take their arsenite oxidizing Ectothiorhodospira-like isolate to transposon mutagenesis and look for mutants that lose the ability to oxidize arsenite.
Changing temperatures might influence the composition of the microbial community. However, we’ll learn next week that bacteria typically have a pretty wide temperature growth range but that increasing temperature (to a point) does allow them to grow faster.
We think of the hydrocarbons as a pollutant but for some of the bacterial community they’re another food source. The bacteria that can utilize the hydrocarbons have an advantage (more food) over the ones that can’t use the hydrocarbons..
Good question. You’d usually want to process samples quickly but people often go to Antarctica for a field season (3 months or more) and don’t process their samples until they return to their home lab. Prolonged storage could definitely alter their results. Not all bacteria will survive equally well sealed in a tube at 4-8 C.
R2A is a popular choice in this work because it supports the growth of a lot of different bacteria. Look up the recipe if you haven’t already (I think I asked you to for one of teh pre-labs!).
No matter what media we choose we’ll bias our experiment towards some bacteria and against others. We just have to accept and acknowledge this!
Correct. TSA is much richer than R2A which is not a good thing when you’re trying to cultivate a wide diversity of bacteria. The weeds grow too quickly on TSA!
This is much too detailed. We don’t need to jump into how the chemical analyses work. We want to focus instead on what the authors chose to do and to measure, why they made these choices and what they learn from them.
Don’t get bogged down in technical details. Focus instead on what they’re measuring and why they’re measuring it. We don’t know enough to dive into the technical details of the chemistry methods.
This is interesting and relates to chapter 5 where we saw that bacteria can have really wide temperature growth ranges. It’s also interesting that these Antarctic bacteria grow best at temperatures they rarely see in their natural environment!
They’ll test each isolate separately and temperature should affect the results much. They’re already shown that most of their isolates grow at higher temperatures than are typical in the Antarctic.
There is no perfect media. R2A and M9 are typical choices for this kind of work. R2A is complex and supports the growth of a wide variety of bacteria (why we use it in lab). The M9 is defined and allows them to supply a single carbon source such as phenanthrene. The M9 also gives a “cleaner” background than R2A/R2B for doing extractions and GC-MS or HPLC analysis.
Diesel usually contains phenanthrene, which is why the authors used diesel-contaminated soils to isolate phenanthrene degrading bacteria.
Soil doesn’t have a defined chemical structure – it contains a mixture of components so we can’t really say anything about the “structure” of diesel contaminated soil. However, we can say that such soil is likely to contain phenanthrene and other PAHs.
The key here is that the different substrate utilization patterns allow them to categorize their isolates into groups. We’ll do something similar in lab 5.
Remember that these isolates are members of different bacterial genera – they should behave differently. Think about the variation in height or hair or skin color we see in our classroom even though we’re all members of the same species! The bacteria in this paper should be equally diverse!
What other data did they collect about these three isolates? The answer to your question is that they probably looked at all the data and not just the phenanthrene adherence to make this decision.
It’s notable that all 3 isolates can grow on phenanthrene but that the diesel, which contains a mixture of hydrocarbons, does not support the growth of P. guineae (and might even kill it – would be interesting to test this).
Think about an oil and vinegar salad dressing. When you shake it up it forms an emulsion where the hydrophobic and hydrophilic solutions are well mixed. Bacteria often produce surfactants to allow them to emulsify hydrocarbons so that they can access them.
We might find a relative in our culture collection. Close relatives of all the isolates described in this study have been found in environments similar to our Gulf coat location.
They’re just saying that their collection of isolates is not necessarily representative of the soil microbial community in Antarctica and that an SSMS system might have favored a different subset of the community.
SSMS is a more “soil-like” system compared to putting some buffer, soil, and diesel or phenanthrene in a flask (which is what teh authors did and what we did in lab, except that we used naphthalene instead of phenanthrene).
I like the link to what we are talking about in class! Yes, they could also take up DNA and maintain it, although in oligotrophic areas the microbes are as likely to use the DNA as a food source (lots of C and N and P in DNA).
The key here is that iron is essential but most iron on the planet is not bioavailable – the earth contains an abundance of iron but not in a form that’s easy for organisms to take up/use (it’s typically oxidised, insoluble, etc). Paragraph 7 has more on this issue.
One thing to keep in mind is that the world was not always as it is now. Life emerged on a very different planet, one where iron was really available due to the anoxic and reducing conditions. Today the planet is oxygenated and the chemistry of iron makes it much less bioavailable under these conditions.
Siderophores are a way for bacteria to capture iron and bring it into the cell. Some bacteria make their own, others steal the siderophores of their neighbours!
The only really important thing for us to take from this paragraph is that they use two different media to grow the vibrios – a minimal medium that has limited nutrients and a rich medium that has lots of nutrients.
Everything else is unnecessary detail – important if you want to replicate the experiments but otherwise TMI.
This is another paragraph with a lot of general information n their molecular methods. The big takeaways are that they made directed knockout mutants; they made plasmids to express some Vibrio fisheri genes ( iutA and fhuCDB).
“polymyxin B was added to the cultures to prevent further growth of V. fischeri ES114 while allowing V. harveyi growth. “
They tell us why they added the polymyxin. It was to stop growth (but not kill the Vf). Why would they wan t to do this? They were looking at the effect of something that Vf secreted into the growth media that inhibits Vh. They screened thousands of mutants so this was easier than filtering them which would have taken a lot of time.
Unlikely. Review how these reporters are made. They are on plasmids but produce fluorescence instead of the transcript (transcriptional fusion) of protein (translational fusion) when the gene or operon is induced.
Luckily they did not have to take samples every 15 minutes. the incubator/plate reader they used could be programmed to take the measurements automatically.
They didn’t set out to study iron depletion. That was just what they found was causing the inhibition of Vh by Vf. They probably went into the experiment thinking it was going to be some kind of quorum sensing phenomenon as this is a paper from Bonnie Bassler’s research group and that is what they study.
It helps to look at the figures. In 1A +ES114 is clear = inhibited. The +MJ11 is cloudy like the no addition control = no inhibition/normal growth of Vh.
In 1b the same data are presented again but quantitatively with the black ars being no addition and the grey bars being the addition of ES114 supernatant. Some species are inhibited (Grey bar < black bar, Vh, Pa, Vc) while others (grey = black, Vp, Vv) are not.
It means what it says – they see inhibition of Vh by the Vf supernatant when they do the experiment in minimal medium but not when they grow the bacteria in rich medium (comparison with rich media result is key). Why would the inhibition disappear in rich media?
I would say that the different media are necessary for accuracy. The different media (rich vs nutrient limited) help define the circumstances under which inhibition occurs.
Saying that Vibrio fisheri si known to produce a siderphore… is not really accurate. to say someting “is known” implies that it was known prior to tis study. At this point all they know is that someting in the Vh supernatant inhibits Vh. They don’t know it’s a siderophore yet.
Vibrio fisheri is not using any filtration method!
The authors used filtration to determine the size of the inhibitory substance. This helped them determine whether the inhibitory substance was a protein (would not pass through a 10,000MWCO filter) or not.
The VH would eventually die but remember the staration stress response we studied in class. Bacteria can use that response and many others to survive stressful conditions by “hunkering down” and not growing, but also not dying.
This paragraph deserves some comments – it describes the first steps in how they found the genes that encode production of the “inhibitory substance.”
You don’t need to understand all the details of how they did it – just that they did random mutagenesis and looked for the loss of the ability to inhibit Vh growth.
Ryne, yes, iron chelation removes iron from a liquid. In this case Vf uses aerobactin to capture iron and then takes up the captured iron and uses it to grow. However, as Dayana points out, this has the effect of leaving no iron available for Vh to use for growth.
The iuc genes encode aerobactin production. We’ll see later that Vf has other genes that allow it to take up the aerobactin+Fe complex. Vh and the other bacteria that were inhibited in figure 1 must lack these genes (or have much weaker versions that are out competed by Vf aerobactin).
Yes. In your summary you should describe the data in figure 2 and interpret it in your own words. The key is in figure 2B where the iuc deletions behave the same as the no addition control with no inhibition of Vh growth meaning …
Dayana has it. The key is that the rich medium has so many nutrients that there is, in effect, no competition between Vf and Vh for iron in rich medium.
Producing aerobactin is costly so Vf only produces it when iron is scarce (minimal medium).
This paper doesn’t address whether siderophores complex other cations but they may.
This looks like the trp operon in some ways – the product of the operon (in this case intracellular iron, raher than aerobactin) acts as a co-repressor of the operon turning off it’s transcription once there is enough iron in the cell.
Exactly. These types of experiment are really important for figuring out gene regulation. The use of fluorescent reporters like mVenus, GFP etc make it much easier to detect transcription or translation than measuring the mRNA or protein directly.
You’re on the right track but need to be more precise.
In figure 4A deleting luxT, yebK, fre or glpK reduces siderophore production which implies that those 4 genes play some knid of role in siderophore production – either by acting as transcriptional activators or supplying a component necessary for making the siderophore. The fifth deletion (glpF) produces the same level of siderophore as the wild type, suggesting that it is not involved in siderophore production (or there’s another gene in the genome that fulfils the same function).
Try again interpreting panels C-D (everyone, not just Makayla!)
In this case it means they did the test in strains with the exact same plasmid – one where they added arabinose to the culture to induce expression, the other with no added arabinose.
The key here is to note the genes that Vf, Vp, & Vv share and those that are found only ini Vf AND to link this pattern back to the results in figure 1 (Vp and Vv were not inhibited like Vh – now we know why).
All bacteria are able to use iron. Iron is essential for all living organisms (I think!) because of it’s ubiquity in the catalytic sites of enzymes.
The key here is how various organisms compete for iron when iron is scare enough to limit growth. Siderophores are one solution that many bacteria use. But some bacteria re cheaters and instead of making their own they steal siderophores produced by others.
Read through the earlier parts to find the function of the deleted genes and then think through how the loss of the genes would affect Vp and Vv. Th iuc genes make aerobactin. What do iutA and fhuCDB encode?
[By producing and secreting the siderophore aerobactin, V. fischeri ES114 improves its ability to acquire iron and, therefore, reproduce, while simultaneously denying iron to a competing species, inhibiting its growth.]
Explaining how this is done is the purpose of this paper!
This is one of the big questions in ecology. You need a balance between cheaters and non-cheaters. If everyone cheats, they’ll all starve for iron!
It’s similar to the dilemma lytic phage face where they need to infect and kill hosts to replicate but they don’t want to be too good at it or they’ll kill all the hosts and die out themselves as well.
This is also part of the equation as Vf has to devote resources to aerobactin production that the cheaters don’t but if we think about cells in a 3D environment the producers may have an advantage in taking up the aerobactin-Fe complexes just because they are at the high end of the aerobactin concentration gradient.
Agreed – it is not a simple system. It’s probably better to be a producer as you are guaranteed siderophores when you need them. if you’re a cheater you’re relying on others so a siderophore supply is not guaranteed.
This is interesting as I would have thought that one way the Vf might benefit from the symbiosis would be through supply of nutrients like iron. It’s good to be wrong now and then!
They’re trying to puzzle out why a mutation that prevents utilization of glycerol as a carbon source would reduce aerobactin expression. They suggest that when GlpK is inactivated the bacterium has to switch to a new carbon source – amino acids. Using the amino acids for energy & growth means that they’re not available to make the protein and enzymes that make aerobactin.
This is a stretch based on their experimental data but it’s reasonable if you think about bacterial metabolism. The experiments to examine this explanation/hypothesis would merit another research paper!
They’re not saying that vents are a source of deep sea nitrate. They’re saying that as as organic matter decomposed as it sinks through the water column the ammonia that is released is converted to nitrate and that nitrate is what makes up the deep sea nitrate reservoir.
Bacteria that don’t require oxygen or sulfide can make a living in this zone. Previous studies have detected nitrification (ammonia -> nitrite/nitrate) and anammox (ammonia + nitrite -> N2) in this zome.
It’s especially important in marine ecosystems because it removes biologically available nitrogen from ecosystems that are already often nitrogen limited.
Good question. In general it is true that oxygen decrease with depth as there is no photosynthetic oxygen production to offset the consumption of oxygen during respiration of organic carbon. We’ll have to pay attention to the data and see if oxygen goes to zero in the OMZ or if there’s enough to allow aerobic ammonia oxidation.
Does anyone else find it funny/strange to learn that water layers don’t mix? These layers are not unusual in coastal systems where freshwater enters the sea.
Storms are one way that mixing of the layers occurs.
Focus here on the data they collect rather than how they collect it. Here is a picture of the CTD-Rosette. The white object is the CTD sensor. The grey cylinders are the go-flo bottles. You lower the rosette with the bottles open and shut them at the depth you want to sample.
It may help to write out the anammox and aerobic ammonium oxidation reactions to see why labeling ammonium or nitrite with 15N can tell how the nitrogen is being metabolized.
CARD-FISH is an adaptation of FISH that amplifies the FISH signal. It’s good for natural samples with low abundance or activity which is why they use it in this study.
The CTD profiles gives us an idea if we’re lookign at a single body of water or if there are “layers” of water that might have different chemistry and biology.
This is an analytical chemistry question. We just have to trust that this is the correct way to do this analysis. The copper is being used to reduce the nitrite so that they can measure it as N2.
they have to label it as putative in GenBank because the identification is based solely on sequence homology and they have no direct functional data to say that it is an amoA gene.
The mRNA vs amoA-cell number comparison don’t quite agree. mRNA tells one story, Cells lighting up with the amoA FISH probes tell another where the AOA are more abundant than the AOB. It will be interesting to read how the authors interpret these data.
Try writing out the equations. In (a) they fed in 15NH4. What process would yield 29N2? 30N2? How do you explain the data showing the production of both?
2d is their model for fluxes of ammonium and NOx. Negative values indicate consumption while positive values indicate production. Does the model match up with the mRNA levels?
Think about what could be driving the changes in concentration in the graphs. For example in figure 1b the decrease in O2 is probably due to aerobic respiration but could also be due to aerobic ammonium oxidation (inferred from low concentration of NH4+ and production of No3- (in Fig 1a) as O2 decreases.
This is a good question. This paper is from the “early” days of measuring anammox and archaeal ammonium oxidation in marine systems. There is a lot more information now and we know that different groups dominate in different environments.
We also need to keep on mind that gens, transcripts, and enzymes don’t exist in a 1:1:1 ratio. Also, their per cell rate if nitrification might not be accurate. There are a lot of assumptions in their work.
The key thing here is that when looking only at bacterial nitrification (AOB) they couldn’t account for all of the nitrification they measured. Discovering the crenarchaeal ammonium oxidizers may account for the “missing” nitrification. (discussed in the next paragraph).
Their PCR primers are probably missing some AOA genes. Think about the PCRs we did in lab. Our “universal” 16s rRNA primers failed to amplify the highly conserved 16S rRNA gene from some of our isolates!
It would be interesting to do some lab studies at different oxygen levels to explore this more. They could need more oxygen than is available or the AOB may be better (faster) at taking up the ammonium that the AOA.
Oxygen levels are probably the key. Different enzymes may work better/worse at different oxygen levels. The data in this paper suggest that the AOA are important nitrifiers in the lower oxic zone while the gamma AOB were active in the suboxic zone supplying NO2- to the anammox bacteria.
The sequences help answer whether different groups are active at different depth. It allows more insight into the system than simply measuring concentrations of various nitrogen compounds.
Right, both anammox and nitrification use the 15NH4 with the norite from nitrification used in anammox. These labeling experiments allow us to see that many different processes can go on at the same time. It’s not always an either/or situation.
A lot of oil wells are located in marine systems and hydrocarbon extraction sometimes involves using saline liquids (think fracking). Therefore, understanding if halophilic and halotolerant microbes can degrade hydrocarbons is of interest. If there’s a spill, will the native microbiota be able to remediate the oil?
Microbes will eat/degrade any carbon source they can access. In marine systems they are often carbon limited and will both produce and consume many compounds we might classify as beneficial or harmful but they just classify as a carbon and energy source!
We don’t know. They’ve been found in the species listed in this paragraph but as they say “…little is known…” This paper will add to our knowledge in this understudied area.
Those of you who took BLY 314 will remember that we used MSM to isolate our naphthalene degraders. What kind of medium is MSM? What kind of medium is Luria broth? Why do you think the authors used two different media to isolate a BTEX degrading microbe? Watch the “Culturing Bacteria” video in module 1 if you cannot answer these questions.
It’s interesting that they used a soil DNA extraction kit kit instead of one for isolates given that they had the strain Seminole in culture (curious but not critical – we’ll use a kit for pure cultures in lab today).
I would guess that these were sites they already had samples from or that they had colleagues that could send them soil or DNA samples.
They do all have either “salty” or “oily” attributes (or both) so they’re similar in some way to the environment from which they isolated strain Seminole.
Yes. Also, there might be some bacteria that can degrade the BTEX compounds but can’t grow on them without other nutrients (for example some bacteria may lack the ability to make a particular amino acid).
Look at the figure and the figure title. The figure tells us the NaCl concentrations they used and the figure title tells us they did three replicates.
Ther could be a few reasons for the lag. #1: the initial concentration of 4-HBA could be toxic and strain Seminole may be inhibited initially; #2 – they maintained the strain on benzene but then tested growth on other substrates. Some of the substrates tested may share a common degradation pathway but others 4HBA might not and Seminole may need time (the lag phase) to synthesize the enzymes necessary to switch from growth on benzene to growth on 4-HBA.
Remember in Genetics when you studied the lac operon? You looked at a diauxic growth curve where E. coli grew on a mixture of glucose and lactose. There was a lag during the switch from glucose to lactose. We may be seeing a similar thing here.
Think back to Genetics when you studied gene regulation. Some operons (for example lac and trp) has all the functional genes near one another. Others (for example mal) has genes scattered all over the genome. It’s slightly more complicated and resource intensive to regulate a pathway that has the genes scattered all over the genome (multiple promoters rather than just one, but it probably doesn’t impose a huge cost on the cell.
Those who took microbiology may remember the citrate test where we detected transport and degradation of citrate. All the bacteria we tested in lab contained genes/enzymes for citrate degradation but not all bacteria were citrate positive – Why? Because come of them couldn’t transport citrate into the cell. The could only degrade cytosolic citrate.
The lack of growth on CAT could be because strain Seminole doesn’t have the enzyme to initiate CAT degradation or it may lack a transporter to bring exogenous CAT into the cell.
I liked this too. Often papers assume we know this happens but people new to the field may not know it so it’s great that our authors made it clear that the hydrocarbon breakdown products enter central metabolism.
I’m not quite sure what you mean by overlapping – maybe clarify your question in class on Wednesday??
Note: You’d usually expect all the genes in a pathway to have the same base name (all pob or all pca) but in hydrocarbon degradation there are quite a few shared pathways that were discovered in bits and pieces and at different times so the gene names aren’t always consistent.
It’s interesting that one of the regulatory proteins (PcaR) is upregulated on 4-HBA but the others (PobR, orfs 3920.3921) are not. Usually regulatory proteins are expressed constitutively.
Not exactly. They report that the paa cluster and this hybrid approach to PAA degradation has been found in many nonhalophiles. They’re simply saying that this “hybrid” strategy has now been found in a halophile as well (their strain).
It’s likely that they simply needed a higher yield protein isolation method. Also, some of the expected but undetected proteins were the smaller proteins which can be easier to “lose” in protein isolation protocols.
Understanding PAH/crude oil degradation pathways is certainly important here on the gulf coast. Even under normal conditions (without oil spills) we have a lot of natural hydrocarbon seeps on the seafloor in the Gulf of Mexico.
We’ll see a lot of variation in the location and arrangement of these genes depending on the organism we look at. I’m curious to see whether they are on plasmids or the chromosome in the bacteria we isolated in BLY 314.
It might be interesting to look up the sequences of the degenerate primers and see how they compare (in terms of the number of degenerate nucleotides) to the primers you designed in class.
The primers later in the paragraph are specific to the genes they found.
Similar principles in that we’re separating biological macromolecules by size using electricity.
Agarose gel electrophoresis is much easier but sometimes you need to look for protein, especially if you make an expression clone and need to check that your host is actually making the protein.
This is a good question. Phosphate buffer is almost always used for these assays. It may simply be that it was what was tried first and it worked so there was no reason to change anything. Phosphate buffer is also more similar to the media the cells were grown up in. A consistent environment is best when trying to get maximal enzyme activity.
PAH degradation genes (and virulence and antibiotic resistance genes) are often found on mobile genetic elements. We often think only of plasmids in this context but transposons are also mobile genetic elements. They can move around genomes and from plasmids into genomes and vice versa.
No. All we learn here is that the (translated) initial 267bp PCR product was most similar to a DBF63 dioxygenase. Dibenzofuran is similar in structure to fluorene so the authors decided that they had probably found a fluorene degradation gene and decided to try to fish out the full gene and maybe an operon.
Figure 2 compares the LB126 genes to those from other organisms. It’s important to note that most of these “operons” are incomplete as they come from similar cloning approaches rather than from full genome sequences.
Best to simply look at the table and focus on the main message – which substrates are degraded and which are not degraded. We don’t need to get into the details of the degradation products in this class – we’ll focus on the genetics rater than the biochemistry.
Protein coding genes are much more variable than genes like the 16S rRNA gene. Also, in exercise 4.2 and 4.3 you looked mostly at one genus where we’d expect sequences to be more highly conserved.
The percent yield is the amount of substrate converted to product. Fluorene was oxidized to three different products, one of which accounted for 63%, the second for 29% and the third for 7% of the fluorene.
Note that not all products add up to 100% and some add up to more than 100%. This is to be expected for a number of reasons – error in quantification, undetectable products, incopmplete transformation of the substrate, etc.
The trick is to try to simplify things. You don’t have to put all this information into your summary. Try to distill the information down to substrates where the products match CARDO and substrates where they match DFDO, and products specific to FlnA1A2. Another approach would be to score products as produced by angular dioxygenation or lateral dioxygenation.
It depends on what outcome you want. Oxygenases are used in the synthesis of some drugs because they produce very specific products – for example one stereoisomer rather than the racemic mixture produced by a purely chemical synthesis.
If you’ve ever taken Zyrtec – only the L-isomer is bioactive but I believe that the pill contains a mixture of L & R.
Enzyme “flexibility” is probably desirable for bioremediation but less desirable for pharmaceutical production.
We discussed HGT/LGT in BLY 314. Bacteria generate a lot of genetic diversity through picking up “foreign” DNA in a variety of ways. This makes up for the lack of generation of genetic diversity from sexual reproduction.
Yes. There are a lot of different ways to inactivate specific genes. You probably learned about this type of mutagenesis in ~10 minutes in BLY 302! We’ll talk about various directed/targetted mutagenesis approaches in more detail in class this week.
It would be better but it can be difficult to sterile seawater and keep all the salts in solution. Also, when doing quantitative chemical analyses it is better to use a defined medium where there will be no interfering substances.
Quite possibly as in the lab we try to control everything – give the bacteria a defined medium with known concentrations of nutrients and carbon. Also, in seawater C. indicus is part of a complex community. In the lab we grow by itself.
The PATRIC CGA lessons cover this. PATRIC generates a similar map. It may help to rewatch the video in PATRIC.
The two innermost rings show the GC content (just in slightly different ways). A bacterial species typically has a fairly constant G/C content. Regions where the GC content deviates from that background level are often regions of DNA that were acquired from an external source by HGT.
The two rings with lots of different colors are showing all of the open reading frames (genes). One ring is the genes read in the clockwise direction, the other ring is the genes read in the counterclockwise direction.
Either side of the multicolored rings (and almost invisible) are two rings showing the locations of tRNA, rRNA genes and other small RNAs.
Perhaps. We’ll have to read on and see if they allow P73 to attack a variety of hydrocarbons or if there’s a lot of redundancy/repetition in the genome.
The arrows represent individual genes they found in the genomes. They’ve color coded the genes with the same function. The numbers below them give their position in the genome.
Figure 5 partially answers Summer’s question from earlier. It maps 38 of the hydrocarbon degradation genes to biochemical pathways. One important thing to notice is that even with a complete (closed) genome sequence they did not find genes for all of the steps.
Notice too the gene numbers. These genes are found in different locations in the P73 genome.
The level of HGT varies between bacterial species. Some bacteria are naturally competent (have DNA uptake systems in the cell envelope) and as a result often have more HGT DNA iin their genomes. However, conjugation and transduction also mediate HGT so there’s no easy way to answer your question.
It looks like some of the dioxygenazses may be invovled in later steps of PAH degradation (phthalate dioxygenase and gentisate dioxygenase are probably 2 of the 6)
Much rarer than it used to be as there are now 10,000s of bacterial genome sequences in the databases. It was much easier to find a “first” 10 years ago!
Think about what you’ve learned in the past about transposons. They can certainly influence gene expression (increase or decrease expression depending on where they insert in a genome).
Yes – we can use genomic analysis to identify genes acquired by HGT. Remind me to talk about this in class on Wednesday. You may have discussed this in a previous Microbiology or Genetics class. Others should feel free to chime in if they remember anything about HGT.
You are definitely missing some information, but your inferences are on target. The genome analysis identified some potential PAH degradation genes and they need to do an experiment (or three) to confirm the function of these alleged degradation genes.
We don’t know! Remember that in the results section they simply report their findings. They are simply telling us what they found. Authors don’t address “why” questions in the results section.
Search the accession numbers in the protein database at GenBank here to learn about the different enzymes they list here. We know these numbers are for protein sequences because the names start with a capital letter and are not capitalized. Gene names start with a lower- case letter and are italicized.
We’ll have to see the results of their Blast analysis to see if your assumption is valid. The sequence matches may indicate if the sequences came from bacteria or archaea or both (or they may not be able to determine this). Can you think of a way to determine whether Mono Lake bacteria, archaea, or both contain arrA?
We analyzed this figure in class on Tuesday. Watch the zoom recording if you’re having trouble interpreting the graph. Figures like this make good exam questions.
We analyzed this figure in class on Tuesday. Watch the zoom recording if you’re having trouble interpreting the graph. Figures like this make good exam questions.
The paragraph title tells you why they added hydrogen. We discussed this in class yesterday. Watch the zoom if you have trouble with this paragraph. Basically they are trying to determine what electron donors the Mono Lake microbes can use (and what electron acceptors).
They identified the arsenite oxidase gene from the arsenite utilizing phototroph a few years after this study. At the time this work was done there were only about eight published arsenite oxidase gene sequences which made designing primers that would target a broad spectrum of arsenite oxidase genes challenging.
That’s exactly what happened. Any time you use degenerate primers for a protein coding gene there’s a danger they will amplify the wrong target and give you incorrect/nonspecific results.
I was interested in the use of these microbes in cleaning up mass pollutants in the environment, and the process by which it is done was interesting. Essentially, workers will attempt to create an ideal environment for PAH degrading microbes to flourish in by adding in carbon, nitrogen, and trace amounts of phosphorous. They will also attempt to increase the surface area of the oil using biosurfactants so that the bacteria might “attack” the particles easier.
Well the phosphorus would make sense as it is a key part in cellular respiration, this would stimulate the activity of any microbes. Nitrogen, however, seems like it would be useful towards any phytoplankton that might break down PAH. So it would make sense that these inorganic molecules would enhance PAH breakdown since they are encouraging the growth of the microbes. At least, this is my understanding of it.
I’m not sure about the time needed, but I do know that 180 RPM at room temp ensures that the bacteria grows an disperses throughout the sample evenly. Perhaps the two weeks is simply provides the best results? And we practice much shorter time frames for convenience sake.
Without reading the paper or reading anyone else’s comments, I think I can derive the meaning of the data displayed in the graph. The control group appears to be a medium with no microbes present. The higher rate of Fla degradation in the YMSM plate tells me that the YE raises the degradation potential of R. erythropolis.
Ah, now this seems vaguely familiar…
My question is in regards to homology. This paragraph states that the colony exhibited a 99% homology with Rhodococcus. Is this from random mutations that might occur in growth? And what is considered the cutoff the colony to no longer be homologous?
This figure is rather confusing to me. Having the 100 and 250 concentrations totally degraded and then nearly 50% of the 500 concentration makes sense following the trend. But then the 1000 mg/l only saw 12.6% degraded. I wonder what could cause this? Does an abundance at that level mean that it is lethal to the bacteria? Is there so much in the environment that there is no need to compete over it?
I think its interesting that it not only can be used as an herbicide, but I did some researching and found that it can also be used to degrade herbicides! Im looking to see if it can also be used to degrade pesticides, and if so it can create serious benefits for our environment.
I’m happy to find that Rhodococcus can be used to degrade PAH’s! With oil spills, overuse of pesticides, and general pollution happening every day, it helps to relieve me knowing that we are finding new ways to deal with the constant pollution we are causing.
I’m still curious as to why an increase in Fla concentration resulted in an inverse rate of degradation. I would assume that it is either a lethal amount of Fla, or perhaps that there is so much that the microbes no longer need to “compete?” Though I’m not sure how much scientific basis that last thought has.
Well, at least I Don’t eat seafood!
Still, the process of using microbes to clean up pollutants in an environment seems to be a lot more prevalent than I had initially thought. When I have more free time, I think I’ll look a bit more into the practice and see what other types of uses this can serve!
Perhaps the concentration of carbon in the water might have an impact on its ability to break down these carbon groups? I’d imagine that C is much more available in the air than the sea, and this would explain it if they were more eager to break down any source they could find.
A defined medium would be used in this instance because the researchers were trying to isolate a DNA sequence for a specific strain. Therefor, it would be better to use a medium where all of the ingredients were known to promote the growth of what you want and inhibit any unwanted strain. If this is correct, does that also make this a selective medium?
I’ve had to dust off all of my old chemistry notebooks just for this class! To refresh on the differences between ester and ether for the cell membranes, re-learning redox reactions, looking up what analytical techniques entail, its a good thing I saved these.
I’ve always felt that evolutionary biology was my strong suit, so I am no stranger to phylogenetic trees. That being said, I’ve only worked with larger scale organisms and never anything microscopic. It looks like the basic flow is the same, but genetics play a much bigger role in diversifying microorganisms since phenotypic observations can only go so far.
Does a genes location on the strand carry any significance? I tried looking around but all I can find is discussing gene locations in regards to knowing where to look rather than function. So it seems like the locations isn’t as important. But still, it is interesting to see the location vary!
An ISP is an iron-sulfur protein. And aside from forming the structure of the aromatic deoxygenase, I’m also having a hard time understanding tis specific function, if anyone has any input I would appreciate it.
ISP is iron-sulfer protein. And aside from serving as part of the structure of the aromatic deoxygenase, Im having a hard time understanding tis specific function. If anyone has any input I’d appreciate it!
While I knew that the microbes that live in thermal vents like the ones in Yellowstone and Paoha Island were extremophiles that thrived in hot conditions, I never thought about their nutrient sources (or much of them in hindsight). I never knew these thermal pools were toxic too, and I’m interested in learning more about the metabolic processes of these bacteria as we read the paper!
Well if we assume an extremophile is simply an organism that can survive in conditions that are unfavorable to humans, I would say yes based on the fact that even if PAH’s cause harmful effects after repeated and longer exposures, they still are an environment humans cannot live in long term. So I think it would have basis to say that PAH degrading microbes count as extremophiles.
Well the color of the hotsprings would suggest a different chemical or microbial makeup compared to the others. zAs for why detail was went into for the location and other geological features, I would imagine that all kinds of data is recorded in regards to the location of extremophiles and where they are found. Certain features might hint at certain chemical makeups of the environment, and it never hurts to be precise and meticulous in gathering details!
Did they combine the products so that they could submit the final product to BLAST and compare the clone sequences to one another? Is that something that can be done?
It makes perfect sense that we would see more archaea than bacteria since this is an extreme environment in regards to chemical makeup and heat. In addition to this, I feel like this is one of the easiest papers to read so far! Im not sure if its because we did all of this firsthand in lab or its the third time I’ve read one, though. Maybe a bit of both?
Im having a hard time understanding why the amount of light has any affect on the reduction oo oxidation? Are these bacteria photosynthetic in nature? Does the amount of light have an effect on the electron exchange?
It makes sense to me that MSM plates are commonly used when growing cultures of bacteria. After a bit of digging around, it seems that this is because salt essentially drains all the water out of the microbe if it is overexposed. The textbook tells that a a bacterial cell is majorly made up of water, and the MSM plates used in lab 2 grew too many colonies to count. All of this makes sense, and it seems the minimal salt plates are favorable to grow large amounts of microbes, but not singular colonies.
What is the normal ecological niche of these bacteria and why would they specificly have a specialization to degrade PAHs? How universal is the gene to produce these enzymes? Is the gene on a plasmid and easily exchanged through conjugation or are is it part of the chromosomal DNA?
Where did you read that PAHs expedite growth and increase population rate? I suppose that if we are able to isolate these bacteria using napthalene enrichment media that they are able to use the PAH as a metabolic energy source but are they actually growing more efficiently then if they were using another energy source? Perhaps it takes a longer span of time to completely eradicate a polution problem in the real world due to environmental factors (access to the pollutant, other carbon sources for food, pH, etc) that would be controlled for in the lab?
I understand that the BATH assay tests how hydrophobic the bacteria are but what does this tell us about the bacteria in relation to being naphthalene degraders? Is this just a standard diagnostic test or does it have special relevance in this case?
I wouldn’t think this was necessarily to “characterize” the naphthalene degraders but nowadays you can’t really describe anything without molecular analysis, right? And knowing what you are dealing with in relation to other genomes helps us give a name to what we a working with or possibly even describe a new species.
All the specimens showed sufficient growth compared to the (positive?) control which was simply stated as a physicochemical degradation but they did not describe what process exactly they used as the control.
The cloudy white substance is actually the emulsion of two liquids that usually do not mix. It is what they are measuring in the emulsification test. An emulsion may look kind of soap bubbly to opaque white like mayonnaise. They measure the size of the emulsion compared to the total volume height to get the emulsification activity ratio.
I believe the more effective degraders (even if they have lower growth rate) are better emulsifiers and have higher hydrophobicity. But just because you can degrade it faster doesn’t mean you also are the most efficient at turning around the carbon into biomass. That being said, I think it would be interesting to compare the size of these particular species and see if volume to surface area does play a role.
Has a study been done to see if asphalt and resin do inhibit gram positive growth or was this an assumption on their part? Obviously some gram positives are capable of naphthalene degrading but to make the statement that gram negatives “better tolerate and uptake PAH” than gram pos due to cell wall structure shouldn’t a more specific comparative expirement be done? (I read the Tebyanian paper cited here and saw no refernces to cell wall differences and in fact the conclusion to this paper was that their gram positive Tsukamurella was the better degrader of hexadecane which is not even a PAH)
Perhaps the gram positive’s thick peptidoglycan membrane (instead of the gram negative’s two membranes) allows it to be more adaptable to a constantly changing environment?
This research, and most of the cited papers, deal with marine bacteria. Do the traits of being bioemulsifier/biosurfactant producers and optimal growth at 400 ppm apply to soil bacteria as well?
This is the first time I’ve seen substituted PAHs mentioned. Are substituted PAHs more difficult to degrade/use due to the constituents of the side chains or is it relative the size of the overall molecule?
Are we expecting the enzymes–and genetic regulation–used for the degradation of these PAHs found in a marine species to be significantly different from those found in terrestrial bacterial species?
They used indole crystals as an indicator to differentiate the E. coli that contained the Cycloclasticus genes for naphthalene degradation, just like we do in lab. E. coli are not normally PAH degraders (I assume) and so they can be confident those expressing the indigo are carrying the genes of interest.
The E. coli is grown on agar supplemented with antibiotics to select for only the bacteria with recombinant genes. This is possible because the researchers included an antibiotic resistance selection marker in the formation of their plasmid. Standard practice in this type of experiment but was not explained.
Overall the percent identity matches are low, coupled with the divergence from the nah-nod-dox-pah genes mentioned later, suggests the Cycloclasticus genes are not closely related to other known genes for pah degradation.
The way the previous paragraph was written is a little misleading. They did have a gene for the small subunit, it just was not located in the “normal” flanking region paired with the large subunit gene. If you look at table 3 and the dendrogram again you will see phnA2 was what they were designating the small subunit gene.
How would we test whether the proteins are expressed constitutively or coordinately? By growing the strain on substrates with and without PAH and then performing an enzyme/activity assay?
So, if I am reading it correctly, the PhnA2 is the beta-subunit partner of PhnA1. Together they make the functional ISP (iron sulfer protein). PhnA1b is only an alpha subunit and is missing its beta subunit and thus should not be functional based on previous research. If it can bind to this other protein’s beta subunit then it could still be useful to the organism. Otherwise, is it just a leftover bit of gene that doesn’t do anything?
These bacteria use As (III) as the electron donor instead of water and so the waste product is As (V) instead of oxygen. They still perform photosynthesis as they still convert light energy into chemical energy, there are just different chemical reactants and products.
I also read that anoxygenic photosynthesizers use a different type of chlorophyll than oxygenic photosynthesizers.
Are the Archaea found in these biofilms part of the As (III)/As (V) biochemical cycle or are they serving some other function in the biofilm community?
If all of the aoxB clones failed to show similarity to anything in Genbank, does this suggest a problem with the primers used or might it have been a novel gene sequence not previously described/uploaded to GenBank?
When reduction of arsenate is driven by H2 or sulfide, with their higher reduction potential, more energy is released that can drive carbon assimilation into biomass. In the light incubation the biofilm seems to be busy oxidizing arsenite which doesn’t give off energy to drive assimilation. Maybe another process was assimilating the carbon that didn’t have to do with the arsenic oxyanions?
Since PAHs expedite growth of bacteria, thus increasing the population rate, why is there such an excess of these pollutants? Is it due to the fact that the enzymes present, even in larger populations of bacteria, are supersaturated with the pollutants – meaning too much to degrade entirely? Or are there instances where the bacteria is enough to eradicate the problem?
Is 94 degrees Centigrade not high enough to denature the DNA, protein and enzymes of the bacteria? What is the optimal temperature range for the bacteria?
Can a reliable trend and correlation be determined from the data? It is leaning toward a trend of positive correlation, as the E24% increases, the BATH% increases as well. However, there are many that do not follow this pattern linearly.
why are two-ringed aromatic compounds easier to decompose? Is it due to more steric hinderance in the electron shells making the bonds easier to break?
In general, how much do the bacterial populations from marine environments vary? The previous paragraph discussed sediments from Tunisia, how different are they from the populations else where, i.e. are specific/unique species in certain locations?
As stated in the article there are 54 different bacteria, what are the specific advantages for these bacteria? Are they not all competing for the same nutrients or is there so much contamination that allows this level of diversity with out worrying about competition?
If seafood contaminated with PAHs were treated with PAH degrading bacteria and sterilized, will harm still be inflicted on human consumption of the newly sterilized seafood?
why bacteriophages? Is there less variablity and more guaranteed that transduction will occur? Is transformation and conjugation less likely to occur or not as easy to measure if it were to occur?
Is this gene required for all naphthalene degrading bacteria? Are there naphthalene degrading bactiera that do not have the ability to oxidize indole to indigo?
At what percentage is considered too low to be considered a different function of the gene? How much can be based on variation and genetic diversification versus random coincidence in amino acids?
Why does it appear that plasmids are more likely a source for PAH degradation genes as opposed to the chromosomes, when it comes to the species found in the environment? Is that specific for this enivornment or PAH degradation as a whole?
Could it be possible the the energy accumulated in the photosystems from the photons of light take a while to de-energize during the dark cycle, hence why it is a longer period?
For a practical sense of research, how useful is this. Due to the fact that different environments will have different bacterial systems and biofilm formations?
When it states exploring what electron donors, is the environment exposed to different compounds or will these temperatures or time changes cause a pronounced change in the electron donors itself?
It is amazing what you can learn from just reading this first paragraph so far. I knew, from what I have learned in organic chemistry, that cyclic aromatic compounds could either be harmful or good but I never know that they affected us this bad. I never really thought about it on such a small scale that goes all the way done to the make of different pollutants.
If the organisms needed to resolve matters like oil spills and other chemical pollutants, why does it take so long to resolve the matter? Also have there been changes to procedures to help ensure that problems like this do not happen or are taken care of in a timely manner?
I would like to know what emulsification activity is. I did not quite understand what it was or the purpose it served when I read the materials and methods section.
Are there naphthalene-degrading bacteria in the gulf coast near the oil rigs that produce biosurfactant and bioemulsifier? Are there more now in the gulf coast compared to before the big oil spill?
Are naphthalene-degrading bacteria used more than phenanthrene-degrading bacterial in most labs? Would phenanthrene-degrading bacteria have an effect on culture samples in lab, depending on if there are certain oils present?
After looking in the genus Sphingomonas I found that thy are all gram-negative and rod-shaped. They are either yellow or off-white in color and have about 37 species.
Are there specific bacteria in the genus Ectothiorhodospira that they are looking for or they just interested in the fact that they are purple sulfur bacteria?
While doing some research on Ectothiorhodospiraceae I found that they belong to an order called Chromatiales. In that order there is a genus called Halothiobacillus that is not photosynthetic.
This paragraph does a really good job at setting up the current issues and importance of Antartica to our global climate. Selfishly, I would love to go and explore the wildlife that I’m only able to see on National Geographic, however, if the cost is this high should tourism even be allowed? There must be some sort of regulation in the future until we are able to leave less of a footprint while visiting as tourists.
It is actually really amazing to me that a place such as Antartica would be so well equipped with NAH degrading bacteria species since their would typically not be a huge use for them. That being said it seems like it is a race against time because the scientists themselves must be using some sort of fuel, most likely diesel, so a really strong NAH degrading bacteria must be found before it is too late. Is biostimulation already in use?
While reading this the question came to mind, does this area have a deep layer of permafrost (I assume it does)? If so are the PAH degrading bacteria located in it? Or is there an active layer of it that the bacteria are located in?
I wonder how they quantified areas as “diesel fuel-exposed” or not. Did they pick soil areas that were in close proximity to ports or roads? Also, I was wondering why they only chose three strains from the 53 that were PAH metabolizing?
I was able to understand the explanation on the results but got really confused when looking at the figures. I am mostly confused as to what is going on in the graphs in part G and H. I can see how D43FB has the highest degradation in figure A but is then outdone by PAO1 which I think is just something to compare it to in the lab, and probably a more common and well-researched bacteria.
It seems unprecedented that a bacteria could be so well adapted to not only be a great degrader but to resist heavy metals. This must have been extremely encouraging for the scientists. I wonder if prior to the experiment they had any idea that they would find this bacteria in the soil? Had it been discovered or researched before this project?
I was wondering if this work was followed up by any bioremediation techniques? Is there not enough information in this research and previous research to support bioremediation using D43FB? I know that they are all aware of the current issues and the urgent need for bioremediation but do more tests need to be done before they are allowed to use bioremediation in Antarctic regions?
The more I read about the soil collected, the more I am amazed at the diversity of the soil in this region of the world. When I think of Antarctica the last thing I think about are what types of bacteria are in the soil. It is very interesting to wonder how some types of bacteria are found very far away from each other in similar conditions.
I’m wondering if bioremediation of the sea floor could work the same way as bioremediation in the arctic. I am also curious about if there were any bioremediation efforts immediately after the Deepwater Horizon spill.
I’m wondering how the scientists were first able to find these various deep sea environments. Also how are they able to collect samples from such places that are over a mile deep in the water and are such harsh environments (pressure, temp, etc).
I was wondering about the change in temperature. Since it said the soil at lower depth was found up to almost 50 degrees, storing it all at +4 C, wouldn’t that interfere with the microorganisms growing at depth. Or are they irrelevant to the study?
Just out of curiosity, I was wondering how long it might have taken these oil droplets to get into a soil sample like the one that is pictured? It seems like it would take a very long time to not only filter down through over a mile of water and then into the soil.
I’m also wondering about the ability to degrade PHE just like in the arctic. There it was due to increased human activity. Is this area of the ocean somewhere where there is frequent traffic of ships using diesel fuel? Or potentially near an area where there is a lot of offshore drilling rigs?
Since some bacterial taxa have been identified that have the ability to degrade PAHs will there be similar bioremediation efforts such as the efforts used in the arctic regions? I was also wondering if deep-sea sediments were the only place where such bacteria are found. It seems like heavily commercialized areas such as marinas could benefit from the same bacteria in much more accessible and shallow waters.
In our last paper it was interesting to me that the same species of bacteria were found on different sides of the globe even though they were found in similar conditions. For them to find the same bacteria in different regions of the sea floor makes more sense to me because the ocean is all interconnected.
It is interesting that they found so many different metabolic activities being used, especially with a compound that is usually thought to be deadly. It goes to show how interesting microorganisms can be.
I was wondering what gives the red coloring to the biofilms in the springs? I have never seen hot springs but when I think of biofilms involving water I think of the green biofilms that are characteristic of a lot of ponds and swamps.
Morgan, I was also wondering why the clones did not show up in GenBank. Maybe they mutated or the isolation process was not exactly right and gave them a different result than what they were hoping.
From reading this I thought the same thing that you did. It seems like natural conditions would favor the reaction occurring for there to typically be a higher concentration of As(V).
Since reduction and oxidations happen in coupled reactions, this graph seems to illustrate that point. I do agree with you though it seems like the reactions take longer in the presence of light.
Like Julie, I looked up more specifically where PAHs come from. For factories that burn coal or where exposure to workers are concerned, or even cities where burning these materials are present, I wonder what the percentage of disease is compared to a population that has minimal exposure. Based on this first paragraph, it would be indicative that disease percentage would be higher in an exposed population versus an unexposed or minimally exposed population. That being said and with research available, I wonder what safety precautions or measures are taken to protect factory workers. Would a certain type of mask suffice? Would burning something like coal introduce PAHs into the air that would then settle on maybe a nearby field that has a food source, which can then be ingested by animals or by us?
Maybe I missed something, but would a LMW or HMW for PAH-degrading microorganisms mean something, as far as use? Would mentioning it pertain to economic cost for production or transport?
I wonder if these enzymes could be isolated or used as a model in some way and then used to degrade PAH. However, if there are no adverse implications to using Rhodococcus, I suppose using them to degrade PAH would be self sustaining and more economical. I would be curious to see how else they would interact with their environment.
Here a 5% NP and 1% of the other PAHs were used, but I wonder what a higher percentage of those sources would do for the experiment? Would it be beneficial or pointless? Also why were alcohols used?
Was the MSM plate used at first to encourage a general type of bacteria to form? In the book, it says that an enrichment culture is used to select for certain microorganisms. Certain media and incubation conditions are also used to select. I suppose the colonies transferred to LB would be selected for. Also, it says that the colony CMGCZ was inolculated in LB and incubated at 30 degrees Celsius and 180 rpm for 48 h. Why was it spun (180 rpm)?
The Fla degraded quickly in YMSM, but I wonder why the MSM took so long? And why did it quicken after the fifth day? Why would removing Fla change this?
Does this mean that if CMGCZ was commercially used one day that it would not be able to handle large amounts of Fla or would CMGCZ have to be added in more abundance? Perhaps the goal isn’t to just use CMGCZ but rather just use it as a model that can later be developed to handle more Fla.
I’m interested to know how Rhodococcus or even CMGCZ would interact with other things or organisms in the environment. Would the Rhodococcus be released to do the job of degrading PAHs and then be done or would it have any negative effects on the environment? Perhaps the perk of PAH degrading would outweigh the consequences, whatever they might be, of not having a degrader.
I was researching PAHs a little and found that LMW PAHs are acutely toxic, while the HMW PAHs are mutagenic and carcinogenic. This research seems promising in that LMW PAH degraders are more successful in research, since LMW PAHs are an immediate concern, especially to aquatic organisms.
I found that PAHs are particularly a problem when mixed in with soil where they build up and are not easily obtained once there; aquatic environments seem even worse off. Here we have the mention of oils. If the oil were to spill in the soil, or even in an aquatic environment, like in the BP oil spill, how would this increased difficulty of PAH breakdown be addressed? Perhaps future experiments would show the promising PAH degraders in environments such as these.
In a couple other studies I’ve look at, fungi and bacteria like Rhodococcus were used to degrade PAHs, and while the bacteria did well in degrading LMW PAHs, the HMW PAHs proved to be the problem. The bacteria seem to only partially oxidize larger rings. With the fungi, PAH degradation is not their ideal source of carbon, but they will degrade the PAHs to a detoxified metabolite. Perhaps a mixture of both bacteria and fungi could help solve the problem? That is, if they’re compatible and not harmful in combination for the environment.
It is interesting that grass fires produce PAHs. If I recall correctly, our local area with Weeks Bay Reserve performs clearing of certain areas using fire. It would be interesting to observe the amounts of PAHs in the nearby waterways, especially the dead end canal areas. Perhaps they take this into account and use something to degrade PAHs?
I was looking into how PAHs were produced, and we know that now they’re mostly produced by industrial activities, but I was interested in how these bacteria came about to degrade PAHs before industrialism or synthetics came into place. Perhaps there was an organic source? As it turns out, PAHs can be produced biogenically by plants and pyrogenically, like the article suggests earlier. Biogenic PAHs are produced by the breakdown of vegetative matter and get trapped in sediments. Interestingly, biogenic PAHs tend to have more rings compared to petrogenic (from industry) rings. Different sources of PAHs also have different ratios to what kind (ex phenanthrene vs anthracene ratio). It was a little unclear as to why the breakdown of vegetative matter would produce PAHs, which would later be utilized by PAH microorganisms. However, the different sources of PAHs and different structures produced by such were interesting to read about.
I wonder what using a mixed, but filtered sample of PAHs from an environment would do? Use the purified ones here, and evaluate results but then use a mixed one from the environment as a step forward from the isolated ones.
It seems that 1-methylnaphthalene has a high conversion rate. What does the methyl substituent contribute towards conversion that the plain Naphthalene lacks? Also, why does placement matter, in comparison to the 2-methylnaphthalene?
pH1a and pH1b had the ability to oxidize indole to indigo. Does this mean that our degraders might have these constructs as well? Or perhaps, how common are these specific constructs in degraders able to oxidize indole to indigo?
I wonder if the phnA4 gene, which has an N terminal region similar to the chloroplast ferrodoxins, are somehow related or came from a common ancestor? Also, it is interesting that P. stutzeri was mentioned here. When I looked it up, it said that it can be found in human spinal fluid, and here, it is mentioned that phnA4 has 47-51% similarity to the NADH-ferredoxin oxidoreductase components. That is interesting that they are so similar and I wonder what the history of this similarity is.
We have plasmid versus no plasmid. I wonder if the Cycloclasticus sp. strain A5, somewhere in it’s history, had a plasmid or fragment of DNA copied into its chromosome, like an episome?
I suppose the difference in location, sequences, and clusters would indicate that different regulations between different microorganisms came into play, even though the basic features of the protein family were conserved.
It is interesting that in E. coli coexpressed alpha and beta subunits were unproductive, but in Cycloclasticus sp. strain A5, the opposite was true. It seems here that the close, “contiguous” placement of the alpha and beta subunits with the genes needed for electron carrying are placed rather specifically. This seems efficient and concise.
Here it mentions that the three dimensional structure of the NP dioxygenase of Pseudomonas sp. strain NCIB 9816-4 has a long narrow gorge which allows it to be able to break down the PAHs that it does. This is an interesting mental picture of what is going on with some of these bacteria. It then mentions conserved residues and divergent ones in the channels in Cycloclasticus sp. strain A5. The diversity of Cycloclasticus sp. strain A5 may contribute to its ability to break down various PAHs. Earlier, it mentioned that Cycloclasticus sp. strain A5 did not have a plasmid, but I wonder if this genetic information coding for this ability would be able to be excised (Ex CRISPR) and placed into a plasmid (like from some of the other bacteria mentioned here) in order to try and get other bacteria to take it up? Or at this point, we wouldn’t even need a plasmid; rather just CRISPR and specific placement of genes.
If primers were the issue, perhaps they could study other arsenic using organisms, use transcriptomics or proteomics to help identify genes and usage of such; and by doing all of this, perhaps they could come up with another primer that would be a success? Perhaps that is too naive to think, though.
I did a little research, and like the PAH degrader investigations, researching arsenic utilization would also be useful to us in breaking down toxins. Orchards, due to fertilizers and commercial treatments, tend to have a lot of arsenic, as well as other inorganic compounds present in the soil. Knowing this, it would be interesting to not only study arsenic degredation but how that metabolism is affected by the presence of other inorganic compounds.
Did they choose those specific times for sampling because of environmental changes (hot environment, cold environment)? Even so, the water is noted to stay pretty constant, near 45 degrees Celsius. Would a more consistent study use a more consistent spacing in time, or even sample once, every month, during the same time of day?
This is an interesting detail. Perhaps the toothbrush was used just to separate the microbes, while the spatula was used to keep the DNA intact. I am not sure, though!
Perhaps there is a difference of initial rate of change from oxidation and reduction states in the light vs dark reactions because, like we discussed in class, there was a selection for a characteristic that normally would have been selected against (fitness wise) in a light environment; but it became advantageous for fitness in the dark. I would be curious to see how many generations passed, the rate of change for the population, and a proteomics or transcriptomic study to compare the differences between the microbes in the light and dark.
I would think that they could use a primer from a known organism that does have the aoxB-independent mechanism and use that as a primer for the aerobic As (III) oxidation activity. I am surprised that they did not do this, but perhaps that just gives them a reason to do another study.
I may have missed this but does the light driven As(III) oxidation have such a narrow range for temperature at 50 degrees Celsius because that is maybe what the temperature would be in its natural environment?
Perhaps the temperature range is so wide in the dark because the presence of physiologically different anaerobes are the more important factor here. Maybe this allowed the microbial population that does light driven As(III) oxidation and dark As(v) reduction to produce energy day and night without interruption.
I think they just wanted to see what electron donors best fit their studied organism and avoid bias based on what they suspected. It’s probably just for clarification an confirmation.
Were they expecting to find that chemoheterotrophy drove As(V) reduction? Do these results seem to suggest that chemolithotrophy or photolithotrophy might be more responsible?
With PAHs having harmful effects on the respiratory system, I wonder if there’s a difference in the amount of exposure to PAHs between cigarettes and vapes not only for those who smoke, but also for those who inhale the smoke secondhand.
I find it surprising that the biogeochemical cycle of arsenic is not as studied what with arsenic being found in high levels of groundwater and it being highly toxic and carcinogenic.
So microbial degradation will remove the toxic PAH compounds as well as oil fractions from the effected environment? Do the physicochemical treatment regimes that we currently use to clean oil spills contain PAH compounds?
Could the factories involved in the pollution of the water supply implement some type of prevention plan that eliminates PAH’s before they reach the environment? If the conditions needed for a specific bacteria, that could eliminate the PAH’s, was found they could introduce the bacteria in a controlled environment before it reaches the outside and is subject to unpredictable conditions.
The filter paper confuses me. Why would they add the paper to the medium? Was the purpose of this to possibly separate larger bacteria from bacteria that could fit through the filter paper?
It says that a phylogenetic tree was drawn based on the 16S rRNA gene sequences but were there any sequences that did not fit? What happens to sequences that are new and how are they placed?
Was the method used here the same method as what we did in lab 7? Initially reading this I did not fully understand what it was saying but after doing it, it makes a lot more sense.
Since ZX4 only oxidized 55 of the 95 carbon sources why did they not continue testing with other bacteria that could possibly oxidize the other 40 carbon sources?
Since the strain was able to degrade other aromatic compounds would this be a good indicator that it should be studied more and used in more areas for clean up?
Could the information discussed in this paper benefit us somehow medically? Could there be some type of use for these bacteria against arsenic poisoning?
What was the reason for the large gap in sampling times? Also they kept samples for several months, how many times would they need to repeat experiments?
As we discussed in lab, normally you would expect to find a largely diverse environment full of various species, but with this environment being more harsh there are less species. I find it more interesting that Ectothirohodospira Sp. to be so dominant.
I am a little confused about how the chemistry of the spring waters and the shallowness relate? I may be missing something or not understanding completely.
If the dark As(V) and the light As(III) both have optimal activities at the same temperature would this also indicate that a microbe is acting in both light and dark or is it just adaptation by different microbes.
The Ectothiorhodospira sp. have obviously adapted extremely well in this environment. Do you think having the arsenite efflux gene (arsB) added in this adaptation?
PAH is a toxic organic compound which means Polycyclic aromatic hydrocarbons. Microbial degradation studies suggest that PAH is a successful way of removing toxic substances. What are the enzymes that can degrade PAH and what are the different mechanisms that that can degrade PAH? Degradation is usually initiated by hydroxylation. What are the other enzymes that can initiate degradation? What are the different pathways that PAH are transferred? Are these the intermediates? Does anything happen after the central pathways or is that the end result? Why are the aspects of PAH biodegradation unclear? Why isn’t metabolic pathways seen in marine bacteria?
Fluoranthene is a PAH that is composed of a five member ring. Fluoranthene has been used as a model compound for biodegradation because it is similar to other compounds of environment concerns. Only a few fluoranthene have been isolated from the marine environment. Why only a few? Why are there gaps for the mechanisms for fluoranthene degradation? Why is there little information available for marine degraders?
What is involved with complete genome sequencing and how long does it take to get the results.? Has a complete genome sequence been completed? How many PAH degraders been sequenced? What are the results for the Mycobacterium vanbaalenii PYR-1 genome? What is the purpose of using Delftia sp Cs-1. What is the importance of having a genomic island? (to allow for adaptation or something else?) Genomic analyses of Alteromonas sp SN2 metabolisms PAH. What are the 4 pathways for Polaromonas naphthalenivorans CJ2. Research on the mechanism of PAH degradation is not understood. Why? Why are there so many unknown metabolic pathways?
Celeribacter indicus P73T can degrade a lot of PAHs which includes naphthalene and fluoranthene. Strain P73T is a fluoranthene degrading bacterium; It can degrade naphthalene. P73T is gram-negative, rod shaped and non-motile. What are the genes involved with the metabolism of PAH? Why is it only a possible degrading pathway?
that’s interesting. I would rather take the easier route. I don’t have the hardest route all the time. lol So is it impossible to sample other waters? I know you said you would need a boat or a ship. Has it been done before?
What is the method of Ausubel et al? Why did they isolate samples of P73T from different areas. (P73T from the Indian Ocean and B30 from the Arctic Ocean) What is the significance of 28 degree C? Are they only able to grow at this temp?
What is Solexa paired-end sequencing? Why were there gaps between the assembled P37T scaffolds? Are the gaps necessary for the experiment to work? What is ABI 3730 capillary sequencer? What is Glimmer 3.070? What is island viewer and why was this used to analyze the data?
Why was the gene delection necessary? What is the cre-lox recombination method? Why was E. coli WM3064 used as the donor strain: Why was pJK100 used as the suicide vector? Could another strain be used for the donor strain and the suicide vector?
What is Solexa paired-end sequencing? Why were there gaps between the assembled P37T? What is Glimmer 3.070. What toold are available in the IMG server?
I think the cre-lox recombination is a knock out process. I googled it and this is what I came up with. In this article they are using mice as a study for cancer therapy. This is an interesting article
okay I found what GC-MS means. (Gas chromatography mass spectrometry) and I know what retention time is. It has been awhile for microbiology. I remember now after looking at the definition (the time required for a solute to migrate, or elute, from the column, measured from the instant the sample is injected into the mobile phase stream to the point at which the peak maximum occurs)
The chromosome has the largest bp size when compared to pP73A, pP73B, pP73C, pP73D and pP73E. The chromosome CDS number is higher than the others. In addition, the chromosome is the only one that contains tRNA and sRNA. I noticed that as the size of bp decrease the CDS number decreases, the G + C content decreases, the average CDS size decreases, the codeing density decreases, and CDS assigned to COG decreases.
This one seems interesting to me because the pP73E is the smallest and doesn’t have a lot of functional categories or cellular processes and signaling. I also noticed that it doesn’t have ribosomes, no cell wall and energy is not produced unless I missed something. Could this be because of the lack of ribosomes or because it is so small? pP73D isn’t much bigger than pP73E but it has some of the things that pP73E does not contain according to this schematic. Does anyone have a different interpretation that would explain why?
Were the genes that encode flagella used for this study? if so, why wasn’t any flagella observed and why was the P73 non-motile? I read that there wasn’t any flagella observed so this would explain why it was non-motile, but it doesn’t make sense if they used the genes that encode flagella.
What is protocatechuate? Why were the genes found in the chromosome and in the plasmid? Why were the genes found in the plasmid vs the ones found in the chromosome or genome vs plasmid?
I found the answer to this question: SIGI-HMM is a sequence composition GI prediction method that is part of the Columbo package. This method uses a Hidden Markov Model (HMM) and measures codon usage to identify possible GIs. and HTG I must have missed that it’s in the paper horizontally transferred gene
ATP binding cassette transporters ( ABC transporters) are members of a transport system superfamily that is one of the largest and is possibly one of the oldest families with representatives in all extant phyla from prokaryotes to humans
The main function of plasmids is to carry antibiotic resistant genes and spread them. The other function is to carry those genes which are involved in metabolic activities and they are helpful in digesting the pollutants from the environment
I found the answer to this one: to carry those genes which are involved in metabolic activities and help in digesting the pollutants from the environment. The main function is carry antibiotic resistant genes and spread them
How many of the 138 genes were studied? Were the genes that encode 6 ring hydroxylating dioxygenases and the 8 ring cleaving dioxygenases studied in this experiment? What other catabolic enzymes, transcriptional regulators, and transporters are involved in the degradation pathways?
Because the other dioxygenases can transfer fluoranthene were they not used because they do not belong to the toluene/biphenyl family fluoranthene dioxygenase groupthis an important factor for them to belong to the biphenyl group?
Because the cost are low and there aren’t any secondary waste treatment this sounds like a win-win situation. However, the high potential for in situ or onsite treatments I’m not to sure about that one. I googled it and it said that the main advantage of in situ treatment is that it allows the soil to be treated without being excavated and transported which sounds like a good idea to me because it said it was cost savings, BUT it said they require longer time periods which could be considered a negative if there as a short deadline
That’s great that 40 species have been isolated from the environment. The bad thing is the article said that little is known about whether PAHs degrading performance can be maintained or encouraged by the presence of heavy metals. I suppose lighter metals will not get the job done and I’m assuming that’s why they used the heavier metals.
I googled ryegrass and it said that ryegrass could take up and transport more ametryn from the soil into the plant shoot. Ametryn is a selective herbicide used for killing weeds. I suppose this would be important if you are using it for hydrocarbon-contaminated soils. Also. ryegrass had the maximum value of translocation factor (TF) for ametryn. I saw a study on google that talked about ryegrass, maize, wheat, and alfalfa. Could they use maize instead?
The three objectives let me know what to look for in the next section. They will test the cooper tolerance, characterize the phylotype and expression of PAH-RHD and c23o genes and study the potential of the remediation of PHE-cooper co-contaminated soils. This paper is easier for me to understand. I suppose because it is the second one. I don’t know. I think I am getting the swing of things
Abiotic factors are the non living parts of the environment which include sunlight, temperature, wind, water, soil and naturally occurring events such as storms, fires, and volcanic eruptions
Abiotic factors are the non-living parts of an environment. These include things such as sunlight, temperature, wind, water, soil and naturally occurring events such as storms, fires and volcanic eruptions
Figure 1-B is a phylogenetic tree that shows the common ancestors. Figure 1a appears circular and 1b appears as a rod shape. Sphingomonas is a gram negative aerobic bacteria.
PHE efficiency decreased as mg/L increased as indicated in Fig S1. At 100 mg/L the PHE efficeincy was 100%. At 900 mg/L the PHE efficiency dropped to around 25%
As the mM of copper increased the PHE removal efficiency decreased. At 0 mg of Cu, PHE efficiency was at 100%. At 7.25 mM the Cu mM dropped close to zeo. Previous studies reported a decline in microbial respiration. I this means that cooper stops degradation. The experiment mentioned iron, so does iron help with PAH degradation?
this paragraph is a little confusing to me. So is this the first study that said that copper promote high levels of Cu? That’s what the last sentenced said that’s interesting. I guess they used nickel instead of cooper?
Rye-grass improved Sphinogobium by proving nutrients. A previous study was done that proved that reyegrass enhances soil peroxidase. Maybe ryegrass can be used to increase other plants growth.
yes I did mistype that. I can’t remember what I was thinking but you explained it in class (the graphs) I think I was looking at something on the graphs
It’s not surprising that combustion or oil related anthropogenic activities cause a lot of PAHs. It’s sad because that means that a lot of environmental waste are the cause of these toxins. In a perfect world you would expect the toxins to come from forest fires and natural oil seeps but that is not the case.
I wander why they used gram negative bacteria. I read somewhere that gram positive worked better for chlorinated dioxins. I’m assuming this one does not use chlorinated dioxins.
a nucleotide sequence is a succession of letters that indicate the order forming allele within a DNA (using GACT) or RNA (GACU) molecule. Sequences are usually presented in the 5′ end to the 3′ end. The sense strand is used for DNA.
Protein exist in different dissociated forms such as amino acids, peptides, enzymes hormones and polypepetides. They carry out different functions in the body. Protein analysis is done to determine the quantity, quality, and the biological reactions in the body. Protein analysis is done to learn the concentration of protein on nutritious food. It is also used to learn the quantity of anemia. Protein analysis is a great way to learn what is going on with a patient. It’s also used in times of crime and forensics examinations. I find this part interesting.
That’s a good question. She said they may not be in the order that they are in the methods section. I think the correct order will be in the results section. Someone correct me if I am wrong
good question Lisa. I googled it and I could not find a good definition for this one. I think it has something to do with substrate specificity, but I’m not sure about it either.
In paper 2 Fig 3, P2 and B1 have arrows pointing in both directions. PHE-1 only have arrows pointing to the left. There are large subunits and small units for all of them. There are black and white arrows. In paper 3, the arrows all point to the right. There are also dark gray and light gray arrows which are absent in paper 2. The dark gray arrows indicate the genes involved in the electron transport chain or phthalate degradation. The light gray arrow indicates the genes that are not involved in fluorine oxidation.
I’m not sure about that. They said they were tested but amplification could not be found. I’m thinking that the primers did not work, but then it said they used angular dioxygenase genes (whatever that means)
my confusion here is what is angular dioxygenase. ? I know angular means having angles or sharp corners and dioxygenase is an enzyme that oxidizes a substrate by transferring oxygen from molecular O2 to it , but I couldn’t find a good definition for both words together. Does anybody know?
Now the lectures on passive and active transport makes sense. The large ones need transport proteins. This makes sense to me. When they talked about it in lecture I knew what they were talking about, but this helps me form a “mental picture” of it .
exconjugants means a protozoan just after the separation following conjugation. Protozoan is a single cell animal from the group Protista, such as flagellate, ciliate, amoeba or sporozoan.
I’m not sure what they mean that the rate-determining step is constitutive which means serving to form, compose are make up a unit as a whole. Figure 1 looks almost the same to me except the black circles are slightly lower than the white circles and the mineralized percent increases over time.
it said strain KN65.2 is closely related to the isolate Novosphingobium. I was thinking that they were saying that horizontal gene transfer occurred, but they didn’t mention horizontal gene transfer. I’m assuming that if they thought horizontal gene transfer occurred that they would have mentioned it here.
Only 50% of the wild type was c, so does this mean that carbofuran wasn’t a good choice to degrade the wild-type because only half of the wild type was mineralized?
what I got out of this paragraph is that group IV mutants degraded at a slower rate, very little decomposition, and no growth. Maybe they should try a different gene to see if they obtain the same results
they used cfd operon. I thought the cfd operon was the reason for the reduced mineralization, so I was thinking for future studies could they use a different gene to obtain more mineralization??
I took an economic botany class over the summer & we went over how yeast is not only an important economic and food staple; but also briefly mentioned how it is used experimentally, especially for its enzymatic properties. It’s interesting to be looking into a more detailed account of this.
I find it interesting how PAH-degrading bacteria from that specific location are observed to be more effective. I’m wondering what determines that; environmental factors, bacterial evolution, both, something else?
It’s very interesting that other than the toxicity and lack of Nitrogen & Phosphorus, all sites present survivable living conditions including necessary nutrients. Is this perhaps why PAHs are able to thrive and are found in relatively high concentrations?
Although the isolates were able to be characterized by unique traits, they all had the gram negative rod shape in common in order to be most efficient in the division & nutrient uptake processes. I wonder if the reason behind their unique properties can be pinpointed, whether it be from environmental conditions, special differentiation, or something else?
I am now seeing that the PAH degraders belong to different classes of Proteobacteria, which explains their unique characteristics. I am interested to know further details for the reason behind why these specific PAH degraders are observed.
I like the flow of this paper & how unique morphology of isolates were mentioned first, followed by phylogeny which provided further specification, and finally the specific abilities each isolate provides in PAH degradation which can be connected again with their unique appearance.
I am interested to see how much diversity is found in the hydrocarbon-degrading microbial deep sea community based off of the sample and how they are described.
I believe that the hydrocarbon-degrading bacteria communities (which are to be identified) are present within the sediment, closer to the surface, and as vent fluids which have petroleum-chemicals present are moved upwards through the sediment closer the surface the bacterial communities degrade the petroleum.
I am interested to see if the hydrocarbon degrading communities observed are present in both samples, or if each sample contains communities unique to their environment.
The use of phenanthrene to isolate degrading strains is because the mineralization results prior to SIP incubations showed presence of PHE in the sample. Is it because it was the only hydrocarbon found, it was the most abundant, or something else?
From what I gather, in paragraph one they are using the Carbon isotope C14 to find which PAH results in the highest level of
mineralization upon exposure to and incubation with the isotope in order to determine which PAH/sediment sample to conduct further studies upon.
Because of the fact that it couldn’t be shown whether PAH degradation is done wholly by Cycloclasticus or initiated by Cycloclasticus which was then consumed along with the PAH it was degrading by another organism without further study proves the intricacy involved in these types of experiments. It’s so interesting that this initial experiment could tail off in other various directions!
I believe they chose these PAHs for test incubations based off of the known presence of PAHs in the oceanic environments they’re studying from prior research.
The sample is identified as Cycloclasticus which is also found in shallow marine sediments. The fact that similar genes at different loci are more than likely responsible for both species of the genus Cycloclasticus being able to function at different depths/oceanic conditions is very interesting as would be a separate study on their comparison.
Based off of this introductory passage I immediately wonder what adaptations or functions within these microorganisms moderate selenite mineralization in order for it to not reach toxic levels that would harm the microorganism, if it would.
I am interested to see the specifics of the fnr regulator and it’s ability to produce a selenium mineral product. I also wonder if this product is considered waste or if it is recycled somehow?
I find this method of measuring the rate of selenate reduction to be very interesting, I believe it would be the most efficient method. Although I do wonder how accurate the numbers will be due to filtration & ion chromatography processes.
I believe that perhaps antibiotics were added to the medium to ensure growth success and prevent contamination. I don’t think it would have an effect on growth rate, just success.
pECL1e was chosen for further study because it was able to rapidly reduce Se VI, unlike the clone pLAFR3 which was unable to reduce Se VI. The change in nutrient medium color indicates precipitation of Se VI and confirms reductase activity.
I believe that after observing the 4 sub-clones had complete ORF which contained fnr and ogt genes they wanted to see if these genes were necessary for reductase activity to occur. The results of cells containing fnr amplification (the gene they chose to study) formed red colonies on agar with selenate, confirming that fnr is necessary for activation of reductase activity.
I am interested to know which specific genes are required to express FNR & why they aren’t present in E. coli S17-1 despite selenate reductase activity being inherent to E.coli. Is it solely because the lack of oxygen sensing transcription factors?
After reading this I wonder if these proteins are perhaps part of a selenate reductase mechanism in E. coli K-12 since fnr gene is not present. I would be interested to see a further study observing pathways in strain K-12 compared to S17-1 to confirm whether there are multiple reductases utilized under different conditions or completely different pathways are used.
I have seen news articles discussing marine wildlife being killed off because of toxic bacteria in the water. Is fluoranthene a source of this problem? If not, then what is the environmental concern associated with fluoranthene?
Since C. indicus P73T is a naphthalene degrader, is it possible that we could determine other genes similar to this gene which may degrade naphthalene also? or is that unlikely?
How long does this process take? Is it possible that this could be used in our experiment using NP as the only source of carbon? Once the metabolites are determined, will more experimentation have to be done on the metabolites? If so, what experimentation would be necessary?
If I am understanding correctly, Does this mean that the organisms in the order Rhizobiales have a strong ability to degrade PAH? If so, do we have access to any of this bacteria to determine its PAH degradation efficiency?
Since it syas that no closely homologues were found in the P73T genome for the naphthalene family, does that mean that we cannot compare the two or does that only apply to this “dioxygenases” section of the paper?
How could this information be resourceful to us? I am trying to understand our experimentation a little better. Will we use the gas chromatography – mass spectrometry?
Is it possible that since “P73_0346 is also responsible for the dioxygenation of naphthalene” as stated in paragraph 3, that naphthalene may have the same or very similar degradation pathway?
I looked up bioremediation to find out more about it and it has a high success rate for oil spills. If anyone else is interested in the article, here’s the website:
Is it complicated to test the influence of ryegrass planting on the microbial degradation of organic pollutants? If so, why is that? and if that is the reason, does the reactivity of heavy metals and PAHs make it complicated to test for degradation?
An ex situ system is a system which conserves and maintains samples of living organisms outside their natural habitat, in the form of whole plants, seed, pollen, vegetative propagules, tissue or cell cultures.
I’m assuming (I may be wrong) by your explanation of the temperature stability of the phosphate buffers that the phosphate buffer is used because of that characteristic. Maybe?
“Protein separation by SDS-PAGE can be used to estimate relative molecular mass, to determine the relative abundance of major proteins in a sample, and to determine the distribution of proteins among fractions.”
Fohttps://www.ruf.rice.edu/~bioslabs/studies/sds-page/gellab2.htmlund on:
“Cometabolic biodegradation of environmental pollutants is a non-growth linked biological process catalyzed by microorganisms. During this process, microorganisms use non-specific enzymes to degrade environmental pollutants that do not support microbial growth.”
In the lanes indicating the presence of fluorine, it can be seen that the bands are dark and noticeable. Whereas, the glucose lanes are either light or do not contain any at all.
To better understand what carbofuran is, I looked it up to compare it to the PAHs we’ve previously studied and found that it is a toxic ands aromatic pesticide commonly used in agriculture. It’s aromaticity, similar to PAHs, is a reason it is difficult to degrade.
Just to clarify, they believe that this mcd gene is the gene that is capable of degrading the carbofuran. Correct?
Also, they found that Sphingomonads are good at degrading carbofuran. So why is it that they used the Achromobacter sp. which isn’t from the Sphingomonad family?
Strain KN65.2 is the strain of achromobacter, correct? I got confused and thought it was a strain of carbofuran. In the previous paper’s experiments, they were testing the degraders so I assume that they are testing the predicted degrader here also.
I definitely misinterpreted the part in the introduction where it explains that they captured a strain of Achromobacter. I see here that the strain KN65.2 is a member of the Sphingomonad family.
When the author says no difference was found, is the slight difference between the two not big enough to be identified? Or am i reading the figure wrong? It seems to me that the carbofuran-grown cells mineralized slower than the glucose-grown cells.
Is it’s degradation level the normal level of copper? Or is copper normally at higher levels than this? Because if it’s normally at higher levels than this, that means that the PHE degradation strain wouldn’t be successful, correct?
I’m interested in why they were not able to obtain an authentic PCR product. Perhaps this will be discussed in the results section. Can I take a wild shot in the dark and guess that the primers did not work as intended?
I did some light reading on the ecosystem of the hot springs. I thought that how the two bacterias the chemolithotrophs and cyanobacteria share a symbiotic relationship to keep their environments ideal for each other.
As(V) is being reduced to AS(III) why would also add 2mM of As(iii) into the assay? Am I missing something fundamental or is this to be sure that As(v) is actually being reduced?
Reading through this section was quite confusing. Are they trying to determine if the bacterial strain is a chemoautotroph or a chemolithotroph? Then above they are looking at a phototrophic bacterial strain also. Did I miss something? This method section is unfriendly and I don’t like it.
It is interesting to see that nitrogen and phosphorus can increase the degradation of PAH. I would like to know how this would work in an event of an oil spill where an increase in nitrogen and phosphorus would lead to an algae bloom. Could there be a way to increase PAH degradation without the use of these nutrients?
I would like to know the time it takes to degrade these compounds is it fast-acting or does it take time to break these compounds down. I would assume it would take longer, due to the nature of aromatic rings being extremely stable. With that said is it possible to introduce this species near waterways where runoffs from agriculture is an issue?
It would be interesting if the same set of genes responsible for PAH degradation in a terrestrial environment is the same as that in the marine environment.
I’m interested in seeing what some of the byproducts these bacteria release in the environment and if we could apply this on a large scale for disaster responses.
I am interested in how they are able to catabolize of these large molecules. Do the enzymes that these genes produce target specific sites? Would the enzymes be targeting the ester group? If so what stops them from targeting other organic materials that have ester functionality?
That is interesting that Phn A can have a PAH substrate range, but cannot convert monocyclic aromatic hydrocarbons. I’m interested in the mechanism that drives the conversion of substrates.
I was wondering, does understand whether a gene is located on a plasmid or chromosome play a part in gene regulation or is this just trying to understand things from an evolutionary standpoint?
Since they found other bacteria containing the Rieske fragment within the sample; are these bacteria able to degrade Fla at the same efficiency as Rhodococcus?
This table actually does not make any sense to me. If you’re sequencing the Rieske fragment and it is homologous to other organisms, how do you know for sure that what you have in your sample is actually Rhodococcus, without sequencing another area of the DNA that would identify the species?
I’m interested in the process of extracting the compounds of interest. How does it happen? Could it just be the polarity difference of the chloroform/methanol “pushing” the other compunds out of the bacteria?
Why would they list a genus that is not part of the experiment? Is it to show that this motiff is conserved in other genuses? I would rather be more interested in what other species of Rhodococcus has this motiff and whether or not if the other species can degrade Fla just as efficient or the other PAHs as efficiently.
If they used zone formation technique for the Rhodoccus sp. why would they change it to spray plate techniques instead? are they using different plates or because they are using a different substrate that requires this technique?
Since the PAH dioxygenase gene is only 78-bp long and in Rhodococcus sp. 100 bp the gene would be conserved. But what are the other bacterial strains being tested are they from different genus? How does the bp length compare to Rhodococcus sp. within the same genus? Is only a section of this gene sufficient for PAH degradation?
[Microbial bioremediation, the process of degradation of contaminants by the metabolic activities of microorganisms, is an ecological, economical and safe approach that can be applied to the decontamination of PAHs with minor alteration of the soil (Bamforth and Singleton, 2005).]
The use of microorganisms to resolve issues in the ecosystem has always intrigued me. Instead of searching for the latest technological advance or contraption, simply seeding a contaminated site with a bacteria that is able to reverse the effects of these contaminants by pollutant-degradation is a clever way to resolve this issue.
I also questioned what natural resources Antarctica had to offer. I am surprised to learn that it, indeed, has more to offer than polar bears and ice! Antartica seems to have an already sensitive ecosystem due to its necessarily cold temperatures. In addition to that, the increasing human activity in Antartica has caused a serious threat to the ecosystem. With the global climate at a constant rise, the oil spillages and the storage and utilization of fossil fuels have greatly contributed to the downfall of the Antarctic ecosystem. It is depressing to see the carelessness of humans and how they treat their ecosystem.
The three strains exhibited almost matching growth yields, while S. xenophagum yielded the highest CFU/mL with phenanthrene as the carbon source. The experiment was then repeated using diesel fuel as the energy source, and while S. xenophagum and R. erythropolis showed even more rapid growth results, P. guineae shows no growth yield and was unable to use diesel fuel as an energy source. P. guineae may be more susceptible to the other particles or PAHs that diesel may contain, which may have prohibited its growth. After a quick google search, it is said that diesel contains 40% phenanthrene, which means the 0.2% diesel experiment contain more phenanthrene than the 0.05% phenanthrene experiment. This may be the reason why S. xenophagum and R. erythropolis showed increased growth yields with the diesel.
These Vibrio seem to be the perfect specimen in this experiment. While being free-living, they are also able to grow under identical conditions, making testing much easier and coherent. Any changes made will show a direct effect without any interference between different environments.
Siderophores with the highest affinity for iron are more suited to live in areas where iron levels are low. While some bacteria are incapable of producing siderophore for themselves, they use heterologous siderophores in order to acquire the necessary nutrients. Siderophore-non-producing bacteria, or “cheaters”, are more like “gold-diggers” in my opinion. They use heterologous siderophores in order to meet their necessary needs while without expending further energy required for their synthesis.
I completely agree that the use of varying types of media will allow researchers to further clarify which supplementations will provide the best environment for the observation of V. Fischer and V. harveyi growth. As stated in the previous post, both V. fishery and V. harveyi are able to grow under identical laboratory conditions, making any alterations made easily recognized.
The process of electroporation is incredibly interesting! By inducing a voltage across a cell membrane, it makes it permeable to any introduced genes allowing the cell to uptake any new genes as seen here with the plasmids beings transformed into E. coli. During the pulse of voltage across the membrane, the cell becomes permeable, but after this pulse has stopped, the cell then “closes” with the new genes inside which otherwise would not have been able to permeate the cell.
[These results indicate that the inhibitory substance produced by V. fischeri ES114 accumulates during growth, and moreover, is either regulated by nutrients, that a component present in rich medium masks or destroys the inhibitor, or that V. harveyi can overcome inhibition if the exogenously-supplied culture fluids contain additional nutrients.]
This is why testing with different mediums are necessary to gather the most accurate results possible. V. fischeri is known to produce a siderophore that inhibits the growth V. harveyi. My interpretation of these results show that the environment plays a huge role in how these vibrio species behave and interact with each other. These results provide more answers to how interspecies interactions are either promoting or preventing cohabitation.
[ V. fischeri ES114 releases aerobactin. Thus, when its culture fluids are added to V. harveyi, iron sequestration prevents V. harveyi growth. ]
V. fischeri releases aerobactin which is a bacterial iron chelating siderophore. These siderophores take in and store, or sequester, all the iron in the environment. This prevents the growth of V. harveyi due to the insufficient amounts of iron in the environment caused by V. fischeri’s release of aerobactin. By deleting the first step of aerobactin biosynthesis, iucD, we have found that the fluids from the same V. fischeri, did NOT prevent V. harveyi growth indicating the cause of V. harveyi inhibition is due to the siderophores produced from V. fischeri.
It’s interesting how nature works and balances itself out. If the number of cheaters becomes overwhelming, there will be a lack of iron. As a result, cheaters population will diminish when they reach a certain point as they are constantly taking in public goods and not returning the favor. Their source will diminish, and they will die off due to the lack of their source and the non-cheaters will rebound. It’s like the circle of life.
I completely agree with Makayla. The experiment should be done a few more times in order to prove these results. As with any given data, we want to be sure that it is valid. As aerE is the main reason for aerobactin transport, it is a reasonable explanation to why this is possible.
It is interesting to see the significant amount of total nitrogen loss when there are such a variety of processes that produce nitrogen gas. I understand that nature has its own way of balance itself out; however, under some circumstances this may not always be the case. This a prime example why the loss or lack of one source can cause a chain reaction towards ecosystem function.
I am very interested in seeing the further studies used in order to find which particular organism is perhaps causing this loss of nitrogen in the ecosystem. Seeing that scientists have already found reliable data which indicates similar findings to what they are searching for in different areas shows for significant hope by looking at differences and similarities.
Flow cytometry is being used as a tool to measure the abundance of microbes in the sample taken. It is also used in immunophenotyping as a tool to help diagnose and classify blood cell cancers. It also aids in finding a form of treatment.
In Figure 1, graph B shows how oxygen is depleted as water depth increases. When oxygen is completely depleted, is when we notice an increase in H2S. This is a possible indicator that another source is substituted in as oxygen levels depleted. In the anoxic layer, other sources are used in the absence of oxygen.
I completely agree with your observation of these graphs! In graph a, 14N 15N almost correlates with 15N15N, however in the presence of 15NO2 (b) 15N15N is shown to have completely flatlines while 14N15N proceeded to grow at a stable rate without the competition of 15N15N. It appears that 15N15N is unable to grow when any presence of 14NO2 as seen in graph c.
At deeper depths, living conditions are definitely less sustainable. There are fewer species that are able to live in habitats with lower oxygen content and little to no light source. These species will also be deprived of many food resources due to lack of species diversity.
I found it interesting that the bacteria found were mono culture where as the Archaea found were more complex. After reading the discussion part it seems that the Archaea are mainly present due to the high volume of salt.
With all the advancements in technology and research, why in 2018 are these harmful compounds still so abundant? Why haven’t more steps been taken before to reduce them? What other ways are there besides bio-remediation to reduce these pollutants?
How long did it take to see the results of bio remediation strategy in The Persian Gulf? Is there any way for these areas to become immune to bio remediation to the point that it becomes ineffective?
Is there a specific reason that these three oil contaminated sites were chosen over others? Why did the samples have to be transported on ice versus at room temperature or at a warmer temperature?
Why did they incubate the culture for 7 days? why not more/less days? Is Gas chromatography the most accurate was to calculate the residual naphthalene? Are there other ways to calculate this amount?
It’s interesting that of 54 strains only 18 (less than half) actually had an adequate growth rate to be further studied. What caused the other 36 to not have adequate growth?
I am curious to see how the results from this paper differ from the first paper we read. Why did they anticipate that this strain would be a good model organism? What characteristics lead to this strain being picked?
How did they choose the strains that were chosen to construct the phylogeny tree? I see in figure 1 that it says “based on a distance matrix analysis of the 16rDNA sequences”, but what does that mean? How were the four clusters in the phylogeny tree determined?
I looked up more information on Pseudomonas putida and found that it is a Gram-negative, rod shaped, saprotrophic soil bacterium. It produces small colonies or patches. Sphingomonas is also gram negative and is non spore forming chemoheteretrophic, aerobic bacterium. It forms yellow or off white pigmented colonies. I find it interesting how they clustered differently.
I wonder if there are any other reports or if there are and discussions on doing more reports about meta-cleavage operon genes in S. paucimobilis to further prove this study?
Besides the salicyclate pathway is there another way for the Phenanathrene to be metabolized by the strain? Could another study be done to find another way?
Why is PhnH hydrolase necessary for conversion? Without it would anything happen? I attempted to look up more information about it but I didn’t see much about it.
From what I know about Paoha Island, it formed due to several volcanic eruptions. I’m wondering if that is going to contribute to why these organisms are able to gain energy from such high concentrations of As(V) and As(III).
Are there only red-pigmented biofilms in this area? If not what others are there and are studies done on them? If so are the results different/similar?
Why did they choose August, October, and April specifically to collect samples? Also, Why was there a gap, October to April, where no samples were collected?
I’m curious to see the results of the temperature range experiment for the light and dark incubated tubes. How is the temperature difference going to affect the samples? Also, how are the results going to vary due to light versus lack of light.
After examining this figure it seems that As(III) is the electron donor and As(V) is the electron acceptor. The temperature of As(V) light incubated biofilm has a wide temperature span compared to the other two. It also shows that the As(III) light incubated slurries and t he PHS-1 light incubated strains over lap.
Are chemoautotrophic bacteria the only type of bacteria that carry out dissimilatory As(V) reduction? If not, what other types of bacteria are known to carry that process out?
Besides Hydrogen and Sulfide, what else can serve as an electron donor for the chemoautotrophic growth of diverse arsenate-respiring prokaryotes? If there are other electron donors, why weren’t they used?
Could further studies be performed to see which biofilm microbes were associated with the arrA amplicons? If so, what would those studies consist of? Would it help the data set of this study?
Could a study be performed to show this cyclic phenomenon for permanently anaerobic ecosystems? If so, why has it not been done before? Also, what would be considered a permanent anaerobic ecosystem?
[Hydrocarbon contamination in Antarctica has profound effects that have been shown to reshape the structure of microbial communities as well as affecting the abundance of small invertebrate organisms (Saul et al., 2005; Thompson et al., 2007; Powell et al., 2010).]
I understand how the oil spillage affects microbial communities in the sediment, but how does spillage directly affect the small invertebrate organisms? From an article online, I found out that nematodes and copepods are two important invertebrates native to Antartica that are important for the shuffling of organic matter and nutrients.
[ Interestingly, this method is the best choice in the bioremediation of soils with low indigenous PAH-degrading bacteria (Castiglione et al., 2016). ]
The article states in the first paragraph that hydrocarbons introduced into soil communities promotes rearrangement of microbial communities, and then implies here that Antartica has soils with low indigenous PAH-degrading bacteria. Aren’t the newly flourishing microbes feeding off the the hydrocarbons introduced into the soil? Wouldn’t introducing PAH-degrading bacteria still upset the natural balance of indigenous microbial communities? I understand the point of trying to find a bacteria to help reduce hydrocarbon pollution, but I wonder about what effect that will have on natural populations.
Yes, the 16s rRNA gene was sequenced for the three highest metabolizing strains found in the samples. They isolated them from the previously isolated 53 phenanthrene metabolizing bacteria from the original 350. Since the 16s rRNA gene is highly conserved between all prokaryotes, it is a great gene to use for the identification between prokaryotic species. I think the three species listed at the end of the paragraph are the three they isolated.
I was curious about the M9 minimal media, so I researched it. It is a minimal, salts based medium to use when you want to grow bacteria with a controlled carbon source and other controlled substances. This gives you the ability to control the carbon source to hopefully isolate certain bacterial colonies; however, the nitrogen source is fixed as it has ammonia present in it.
Although the results clearly support S. xenophagum (D43FB) as being the most efficient PAH-degrading bacteria isolated in the samples, I still have my doubts. For starters, there’s the issue of high cadmium levels (see comment above). Additionally, not stated in the article is the issue of the optimal growth temperatures needed for the strain. The strain grows optimally around 28 degrees Celsius, with negligible growth at 4 degrees Celsius. From googling, I found the average temperature in Antartica to be -10 degrees Celsius on the coasts, and -60 degrees Celsius more inland. How well will this species degrade phenanthrene when actually used for bioremediation, and will a small benefit outweigh the potential effects of artificial seeding?
The article states that the diesel-exposed soil sites in Antartica have levels of cadmium between 15-85 mg/Kg, and the D43FB strain can only metabolize around 20% of the initial phenanthrene when in the presence of high cadmium levels. Although the D43FB strain seems to be the best option due to highest levels of phenanthrene/diesel fuel degradation and biofilm/adhesion formations to phenanthrene crystals, how is the metabolism of phenanthrene by the other two strains affected in response to high cadmium levels?
I think the researchers are just saying that the techniques they used may have favored different bacterial strains than ones that would actually grow in the soil’s native conditions. They achieved their goal of isolating PAH degraders, but there may be ones not detected in the laboratory conditions that readily grow in the soil. They are simply stating a limitation to their techniques.
I could be wrong, but I would assume the bacteria would have a greater chance of forming biofilms in the natural environment compared to the laboratory. I think this because bacteria sometimes form biofilms so that they thrive better in harsh environments and can work together to become more resistant to certain conditions. I think attempting to simulate the soil’s conditions would tell us a lot more, especially considering how the D43FB strain had genes encoding for flagellar components, but chemotaxis was not observed in laboratory conditions. This was important in showing how bacteria can turn on/off certain genes according to their environments.
I could not find compounds similar to siderophores, but I did find that some organisms are able to store excess phosphorous inside of the cell by making polyphosphates. Certain anaerobic bacteria are able to consume carbon energy sources by using these polyphosphates as energy rather than oxygen as an electron acceptor.
From researching, I found that there are hundreds of different siderophores that can have different affinities to iron based on their specific properties. They contain two negative oxygen molecules that bind to the the positive ferric iron, and come in the form of hydroxamate, catecholate, and carboxylate functional groups. The oxygen molecules can be replaced with a nitrogen or sulfur molecule, but this decreases affinity. Additionally, the functional groups can act as hexdentate, tetradentate, or bidentate ligands. They go from the highest affinity to lowest affinity for iron in that order.
I was curious about the CAS assay so decided to research it. Chrome azurol S (CAS) and hexadecyltrimethylammonium bromide (HDTMA) are used to make a complex that binds ferric iron. It can be used to quantify siderophore levels in a sample because as siderophores remove ferric iron from the complex, the absorbance of the fluid sample will change. This assay cannot be used to tell you which siderophores are present because since there are hundreds of siderophores with different binding affinities, this would require several assays that are more specific for certain siderophores. The absorbance of the fluid should decrease when more siderophores are present.
I am a little confused with this sentence. Polymyxin B acts as an antibiotic to gram-negative bacteria because it binds to the lipopolysaccharides of the outer membrane causing the permeability of the membrane to change, leading to cell death. Did the antibiotic specifically only target the V. fischeri because the V. harveyi were conjugated with an antibiotic resistance plasmid from treated E. coli? This is what I assume happened based on the first paragraph of the paper, but was a bit confused on that as well.
The takeaway from this paragraph and figure three is that the Aerobactin siderophore produced by V. Fischeri is controlled by a biosynthetic pathway, since it shows negative control of a repressible operon. Under normal conditions when iron is present in sufficient amounts, it acts as a corepressor and binds Fur, which binds DNA and stops the transcription of siderophores. When iron is depleted, the Fur repressor is not active and transcription of aerobactin is turned on.
Figure 3 proves the Fur gene to be responsible for repressing siderophore production. Figure 3A uses a CAS assay to show that as iron concentration increases, siderophore production decreases; this is not seen in the mutant with the removed Fur gene. In Figure 3B, the promoter for the target gene was coupled with mVenus, a fluorescent protein that can be quantified. As iron increased, mVenus was picked up less, meaning that transcription was repressed as iron increased. Again, there is no effect seen on the mutant.
I believe that this was just an example of what the authors stated earlier in the paragraph. It is a cheater strain that is able to utilize various siderophores secreted by other strains but does not incur any costs by producing it’s own. I could not find on the internet whether it lost it’s biosynthetic genes for producing it’s siderophore or simply gained receptor and importer genes.
The aerE gene is still in the cheater strains because they will still need to export the siderophore out of the cytoplasm. Having this gene is beneficial because it allows the siderophore to not build up to toxic levels intracellularly, and it also allows the cheater to cycle the siderophore for more iron. Because of these advantages, these strains are probably selected for at a higher degree since they increase the cheater’s fitness.
Upwellings make ammonia available to bacteria, who can oxidize this to nitrate for energy. This creates areas of low oxygen, so bacteria have to use nitrate or nitrite as terminal electron acceptors. When they do this, they use either denitrification or anammox for energy. Denitrification involves the reduction of nitrate to nitrogen gas. Bacteria only switch to this in anoxic conditions when no ammonia is present because this reduction consumes energy. Anammox involves oxidizing nitrate to nitrogen gas by using nitrite as the terminal electron acceptor when no oxygen is present.
I was curious about the anoxic conditions of the Black Sea, and found that this is mostly due to there being two layers of water in the sea. Rainwater and river water flows into the sea causing the surface of the sea to be composed of freshwater, while the deeper waters are fed by the Aegean Sea and are much saltier. This creates a large density difference, so the two layers do not mix well. This creates highly anoxic conditions in the deeper layers of the sea.
PCR techniques will allow the researchers to quantify the expression of the amoA gene in different nitrifiers. Additionally, PCR of the 16S rRNA gene and phylogenetic analyses will allow for identification and characterization of the bacteria/archaea that they find.
Since 14N is the most abundant isotope in nature, 15N can be added in so that the researchers can use this ratio to determine the uptake and usage of 15N. During anammox, bacteria use ammonia and nitrite to produce nitrogen gas and water. Aerobic ammonia oxidizing bacteria can convert ammonia to nitrite and nitrogen gas; therefore, the aerobic oxidizers produce a nitrite product that the anammox bacteria can use. The researchers will be able to study exactly how this nitrogen is shuffled around between the two by using 15N.
In the suboxic depth, nitrate levels drop as ammonium levels rise. Anammox bacteria also peak in this area as they oxidize ammonia to nitrogen gas using nitrite since there is no oxygen.
I believe that this figure is simulating the conditions of the different depths by making different nitrogen species available. Only (a) shows production of nitrogen gas made fully of 15N when ammonia is available. When NO2 is available (b and c), this is not produced. This means that in the suboxic and anoxic zones where ammonia is being oxidized by anammox bacteria, ammonia is being used and converted to nitrite and nitrate in order to fuel the anammox. When nitrite is already available, this doesn’t happen.
These results state what I interpreted from the graph; that in the suboxic zones where only ammonia is present, anammox is linked to nitrification to replenish nitrite.
I think the conclusions the authors draw are very interesting. Due to anammox rich areas being shown to be linked to nitrification, this gives a more clear picture into marine nitrogen loss. It would be very interesting to replicate these types of experiments in the oceanic OMZs.
Hi, this is a comment.
To get started with moderating, editing, and deleting comments, please visit the Comments screen in the dashboard.
Commenter avatars come from Gravatar.
For what I understood V.harveyi can be considered a “cheater” because it encodes genes for the byosinthesis of amphi-enterobactin which is has a high affinity to siderophore that acquires iron formicrobial systems.
It seems that aerE provides an adventage in the adquisition of iron and also an adventage of a superior growth.
Interesting experiment, it explained very well how the aerobactin works as a inhibitor molecule for V. harveyi and taking in count that this lead to a quantitative investigation the results can be more precise in future experiments.
After reading about FISH I was impressed with the many adventages it has, since it allows the detection of one to three orders of magnitude more bacterial cells in the samples, allows study of the spatial organization of cells and in top of that, cells do not have to be alive.
For what I investigate it seems that is a constant concentration gradient which is obtained by imposing a limited solubility on the compound.
I was also wondering the same thing about the use of icebergs to supply cities with water and it could bring more damage to the environment than it seems. The towing of icebergs can cause a change in water temperatures which could affect the aquatic life of the area creating a chain reaction that would distort the ecosystem of Antarctica.
Taking into account that the alternative of using icebergs as a source of water supply is not very viable due to the alteration that it would cause in the ecosystem, it is possible that these contaminated icebergs can be treated after bioremediation for an emergency case due to Lack of water ?, Is this “fresh water” still drinkable?
I had the same question and I have done some research. It seems that over long periods of time (two years) it can have an impact on the PH levels causing them to decrease. However, taking in count that the waiting time it was not so long it would not make much of a difference. But, I’m still not so sure why they waited two months to do the analysis.
For what I researched the agitation moves the liquid onto the walls of a vessel and it can increase the surface area which would subsequently increase the oxygen concentration allowing the bacteria to grow faster.
Situ bioremediation caught my attention and it seems that there are many methods that can be used. One of them is the biosparging that involves the injection of air under pressure below the water to increase water oxygen concentrations.
yeah that caught my attention too. Foreign organism are forbidden because of the isolates location, meaning that the local organism have not developed natural protection against foreign species.
When they say “we restricted our analyses to their free-living lifestyles using shaking liquid culture for co-incubations”, what kind of specific culture do they mean?.
It is really interesting how this species interact with each other and the fact that V. Fischeri ES114 can use a filtration method of a 10000 MWCO membrane to prevent V. Harveyi from growing.
Can this Iron chelation also affect the amounts of Magnesium if that was the case?
As stated in the introduction, nitrification only have a good performance in the lower oxic zone, this is because one of the main factors that nitrification needs its a good concentration of oxygen and as we go deeper in water this concentrations of oxygens star to decrease making it very difficult to carry out the process in the suboxic and anoxic zones.
(A). the crenarchea cells showed better performance in the suboxic area and very low or non existent performance in the lower oxic and anoxic areas.
(B). BAOB cells and mRNA performed very poorly inall the zones.
(C). YAOB cells in the suboxic zone performed their best during this time but their performance was very low in the lower oxic and anoxic zones. However, mRNA did not have fized performance where it decreased and increased between zones.
Yes, because port activies can provoke a lot of pollution with ships, transport, diesel emision, and loading of products. Also, we can add the risk of a possible oil spill. In conclusion, Mobile bay could have greater pollution due to high activities than other places that are not this active.
Good point, MSM would be a good media to start specially because of what the scientist are looking for which is a bacteria that can tolerate high concentration of salt like the ocean. After, we can exclude those who can’t biodegrade the hydrocarbons in a high salinity enviroment.
MSM media is considered a slective and differential growth media at the same time because it encourages the growth of certainbacteria while decreasing the growth of other kind of bacteria. In the other hand, we have Lurie Broth is a rich mediaum that allows the growth of a different bacteria and it consist of tryptone, yeast and sodium chloride. The reason that they used both its because by having a rich medium it is highly probable that it would grow in that medium but in the MSM there could an exclusion.
I did some investigation about the UltraClean soil DNA kit and I found it that the soil samples are added to a bead beating tube containing beads, lysis solution, bead solution and inhibitor removal solution. The obective is to lyse the microorganisms in the soil by a combination of heat, detergent, and mechanical force against specialized beads.
it is really interesting that seminole was able to use benzene as a carbon and energy source but the situation changed when the bacteria could not dregrade benze at a 4 M NaCl.
I was curious about this topic two and I did some research an catechol can be degradaded by two ways: the meta cleavage pathway or the ortho-cleavage pathway.
I think is whitin the category mentioned, correct me if I’m wrong.
I’m interested in what kind os metabolic reconstruction process is done when using “-omics”.
I look up fluorene and is PAH used to make dyes, plastics and pesticides. Also, one interesting fact that I found is that it has an aromatic odor similar to naphthalene. It is possible that compounds that have similar odors could have similar degrading bacteria, like there could be any correlation?.
interesting that they also have problems identifying the enzymes that govern the initial attack on the PAH degrador as the scientist from the previous paper.
interesting that they are using gas chromatography for the separation of mixtures. we just used column chromatography separation methos which is similar to the gas method, helped me to understand.
Yes, I agree, this article can help us as a guide for our research and give us a perspective about the process in general.
I have the same question about this topic, if I would have to make a guess I will decide that they both are the same.
I was curious about gene transfer so I ended up google in it and it says that is the movement of genetic material between organisms belonging to distinct groups of interbreeding individuals.
I googled Fluoranthene and is a molecule that can be viewed as the fussion of naphthalene and benzene unit connected by the previously said five-membered ring. However, Fluoranthene is a colorless compound.
I did some research and a study says that the Indian Ocean is the second- most polluted in the world which makes it a escenario to do this kind of research.
It is interesting with the results of yeast. Would it have been more efficient to also use yeast with this experiment at least for a comparison?
It makes sense to isolate the PAH-degrading bacteria from that specific location because it they already adapted. So that factor can be ignored.
I find it so interesting on how the PCR technique works. By just going through cycles of different temperatures resulting the the amplification off DNA is very intriguing.
I wonder why they depended on optical density for bacterial growth instead of having it grow on a Petri dish and counting the colonies.
I believe the environmental factors play a huge effect. Even with the previous paper the bacteria that is closest to that region had the best result of being the most effective in PAH degradation. I am assuming that’s the same case for this paper.
I do not understand how this technique relates to the overall goal of degrading the PAHs. Like how is isotope-stabilization a specific characteristic?
Isolating a sample from the electrophoresis gels can help determine morphology through DNA sequencing.
I know we are not supposed to question too much about the details of the technique, but I truly do not understand the treatment of the controls and what is being compared. Or how it contributes to the overall experiment.
I did some research on the neighbor-joining method, because I was curious also. and it does not seem to be the most popular to make phylogenic trees, but it is a quick way to organize a large amount of data. One advantage is that it shows how everything does not evolve at the same rate.
I was curious about the same thing. How is this information will be used to help solve the overall problem? How are they going to take this information and possibly enhance the productivity of the PAH degraders?
It is interesting how there are structural variations of Se(0) particles from different bacteria that are present. I wonder through evolution how that came to be. Like are the systems similar or just the product. Is it due to an ancestor or just environmental pressures over time?
I was curious as to why this bacteria would use this metabolic pathway over others. Why would it use its energy for this in the first place?
The purpose of adding antibiotics to the media to prevent contamination from other strains. The strains they are trying to isolate are resistant to the antibiotic so in theory its the only strain able to produce colonies.
I believe it is more of a confirmation test. If the bacterial strain is so showing it has the ability to degrade selenium, then there must be a transcriptional pathway that gives the bacteria the tools to degrade.
I believe your question is answered in the next paragraph. It has the characteristic ability to reduce Se(VI).
Yes this appears to be the case, showing that the cosmic clone pECL1e has a significant effect in Se(VI) degradation. This also covers any unforeseen chemical reactions that might have contaminated this test.
That what I go from this paragraph. The purpose was to find out how Se(VI) can be degraded and for this bacterial strain it has to live in anaerobic conditions forcing the strain to use Se(VI) to the the sole electron acceptor this in turns needing FRN regulation to become active.
So the overall result is that the environment greatly dictates the metabolic pathways. It uses “oxygen sensing proteins” in order to to decide which regulatory pathway would be most efficient. So when these proteins do not sense oxygen then that is when the FNR regulator becomes active allowing the bacteria to degrade Se(VI).
I was reading up on the mobile chemistry plant pollution in Chickasaw and I was wondering is it possible that there are cancer possibilities through the aromatics and chemical waste. I’m asking for a family member.
What specific characteristics in the bacteria are they searching for?
Could the chloroplast-type ferrodoxins be involved the coordination of more than the two iron atoms or is this strictly for this cluster only?
Wouldn’t hydrolyzing the products make the structures more water soluble, in which help break it down?
If aerobic non-sporulating rhodococci are great contributors to the biodegradation of environmental pollutants, would it be hazardous to introduce more into the environment to help speed up the end of pollution?
Can Rhodococcus be used to degrade other pollutants other than PAH and is there other bacteria that can degrade PAH?
Along with the diluted culture spreads, I was thinking that a concentrated culture could also be spread on a agar plate to compare between the dilute agar plate spreads.
I noticed that the experiment used MSM plates instead of TSA and R2A like we use in our experiments. What exactly are the differences between using MSM plates compared to the others?
If complete degradation of concentrated Fla is the overall goal of the experiment in this section, why not prolong the incubation period? Will it mess up the data?
I’m not quite sure I’m understanding the chart here, but is it basically saying that every relevant match with a percent identity of 86 shows a closer homology to Rhodococcus sp. CMGCZ than the relevant matches that show a percent identity of 84 and 82?
So I’m assuming that LMW PAH has 2-3 ring structures while HMW PAH has 4-5 ring structures. I find it interesting that even though LMW PAH is more favorable than HMW PAH when it comes to degradation, some bacterial strains degrade HMW more readily than LMW, despite ring structure size.
If Nap, Phe and Fla degradation was inhibited on a Ye medium plate, would it have done the same thing if Nap, Phe and Fla were added on a R2A, TSA or MSM plate?
I aslo think that marine bacteria are able to degrade PAH more readily because the environment they live in might be a little bit harsher compared to terrestrial bacteria, for example, saltwater compared to a forest.
I’m also interested in how terrestrial and marine bacteria differentiate between each other when it comes to degrading PAH. If environment plays such a key role in how quickly they degrade it, then we could possibly colonize and utilize them in quicker degradation of harmful substances.
I did not know that PAH affected the surrounding environments and people that live in them to that extent. I think this is very useful in bringing awareness to the situation so ideas can be brainstormed to fix and prevent this problem.
I love how we are able to do these experiments in our lab to see how we could apply this to real world problems. Even though these experiments are a little more in depth than ours, I still enjoy learning and applying techniques to these types of problems.
For the thin layer chromatography, is there a reason why silica gel is used? I’m just curious because i know that you could also use cellulose and aluminum.
I wonder how the results would’ve differed if the use of a different company was used for each experiment or different purities were used instead.
Hey Jennifre. I had the same exact question that you stated. Maybe it could be due to some gene silencing or maybe it’s just expensive to keep doing multiple cloning.Not entirely sure though. Just throwing out some suggestions.
Could the reason why the cis-dihydrodiols of the latter compounds were converted to monohydroxylated compounds due to ring stability and aromaticity? Also, could it be due to activating and deactivating groups that are present in the ting structures or the types of reagents used on them that made them become monohydroxylated?
Isn’t upper pathways how the original compound goes to a intermediate compound that still contains ring structures, but they are unstable and lower pathways are rings that are being cleaved and de-aromatized?
Is the reason why the data for the location of the genes encoding PAH dioxygenase of A5 wasn’t shown because it was irrelevant to the paper or was it too much to be included or too complicated?
I wonder if obtaining biofilms from different extreme environments would have the same affect as the biofilm taken from the Paoha Island.
I was thinking the same thing Tim. This makes me wonder if there are other chemolithotrophs and cyanobacteria that share a symbiotic relationship to help each other and do the same thing when it comes to breaking down arsenic to use for energy.
I wonder if it had mattered whether they chose from these springs or other springs that are around the area if they all contained dissolved sulfide, ammonia, methane and arsenite.
This is very interesting to me. I wonder if they would have the same results that they obtained from this same location either now or last year and whether different steps could be taken with the new technology they possibly have now.
I was wondering the same thing Sarah. To me it would make since that it would find an alternative way to survive if exposed to a darker region than what is was previously in. If this is true, then this is a cool way to examine how adaptation to a new environment plays a role in metabolic pathways.
Hey Lameace, I’m curious as well of whether there is more than one clone and whether this experiment was successful or not. Maybe they could specify that there are more (or just one), but because this was the more dominant arsenic cycler, they concentrated on it more for the sake of the paper.
I was wondering the same thing Jessica. If a sample was taken from a lake that contained less salt than the lake they obtained the biofilm from, would there be a drastic change or a slight change. This is very interesting to me.
Could the temperature ranges from the experiments be due to the environment that they came from (the springs being around 45C and the microbials halting activity at around 50C)?
I agree with your question Jessica. If they already knew that sulfide and natural gases help to drive the reduction to As(V), then why was the experiments incubated with the same electron donors. Maybe they could’ve used a different electron donor to observe the reaction.
If acetate did not give the needed results for the experiment then what did it do? i think that they should include the results of what was observed with the experiment cause I am actually curious as to what happened.
I’ve always associated aromatics with being more good than harmful. I guess because of ‘aroma’ being apart of the name. I knew that their were some that could do some damage, but to the extent that it has caused in The Persian Gulf.
Is bioremediation the only strategy that can efficiently eliminate environmental pollutants?
Is there any particular scientific reason why sediment samples were taken from 12 cm below the surface and seawater samples taken from a depth of 15 cm?
What is a turbidity measurement, and why would you take such a measurement indirectly assess growth curves opposed to something else?
There seems to be a steady increase then a more dramatic decrease.
The isolates are more extensive than what I expected them to be. The phylogenetic tree gives a lot more insight and a better visual.
With this particular source being from 2014, why is this the only method to break down the crude oil?
Is there a cyclic aromatic compound that would be virtually impossible to be decomposed by bacteria?
What is LB agar?
Since the strain was incubated for 48 hours, would substantial growth have no happened within 24 hours? I am assuming that the growth rate is slower.
Phenanthrene concentration had a pretty steady decrease per the graph.
Could this be due to the compound being more easily degradable?
Since the strain was found degrade several other aromatic compounds, is this indicative of phenanthrene being more stable than naphthalene?
Because of the ability to degrade more than one compound would phenanthrene be a better aromatic to use concerning the issue of oil contamination in the gulf?
What is the possible reasoning for being unable to obtain authentic PCR results?
I was absolutely unaware of this side of arsenic. It has always been viewed as a fatally toxic compound.
Since no clones matched any of that in the database then what does that mean? Were new species possibly discovered?
Why weren’t the single clones included in the analysis? By doing that would the results still be reliable?
Is the RFLP type percentage more important than the identity percentage in determining the representative clone?
Why would the rate be less in the dark?
The PHS-1 Strain was consistent with the Arsenite range when it comes to oxidation.
It is extremely interesting to learn the different operations of both arsenite and arsenate, and their respective processes in light and dark.
My species from 314 was Burkholderia (Paraburkholderia, specifically). When I was researching it, what I found concerning PAH degradation only spoke of toluene. It interesting to read about the study of another of its degraders.
The fact that this strain was the first fluoranthene-degrading bacterium to be found with the family of Rhodobacteraceae just goes to show how much of the world had yet to be researched, studied, and discovered.
Are all PAHs about the same in regards to being harmful to the environment or are there some more detrimental than others?
I wonder why the mechanism of PAH degradation has so much unknown about it. Is it really more in depth than the basic/well known chemical reactions?
What is a conjugal donor strain and why was E. coli used?
Was 28 degrees Celsisus the ASM temperature?
Why does the entire genome need to be described?
This must mean that the P73 strain is soluble in water because of the lipids it contains, correct?
How exactly could the P73 strain using gene transfer enhance its PAH degradation ability.
Would the presence of glucose-6-phosphate have made a difference in results?
Could strain B30 also have been acquired via horizontal gene transfer?
What is the first discovered PAH? I’m sure it has been widely studied. Would that not be a good basis for studying the physiology of not just fluoranthene, but other PAHs as well?
Since in situ is low cost can it not be used as a treatment for PAHs?
Does ryegrass contain some type of chemical that makes it helpful in biodegradation?
What does the methylene chloride do in the liquid-liquid extraction?
Why was MSM supplemented with glucose used as a blank control? Or rather,why wouldn’t MSM by itself be sufficient as a blank control?
What could have caused the decrease from 24-48 hrs? Is that just a naturally occurring circumstance in this specific gene?
Planted vs unplanted soil? Is this soil that grew plants and soil that did not?
Since xylE and ahdA1b-1 had consistent results to each other when promoted by Cu(II), would this mean that both genes have similar phylogeny or genetic makeup?
So would planting more plants in rhizospheric soils be a solution to lowering the level of PAH concentration in the environment?
So fluorene degrades differently than the other PAHs?
Why is there not a lot of data concerning fluorene and gram-negative bacteria? Does that mean gram-positive is more associated with this particular PAH?
What does the phosphate buffer do?
Why did the extracts need to be dissolved in acetonitrile? Why couldn’t the dried extracts themselves be used as they are?
What does it mean that no amplification could be obtained? The PCR did not work?
I see that they used a set of 15 primers. So you are not limited to just two primers? More than two can be used at a time?
Is the reason that phenanthrene cannot be used as the sole carbon source because when the strain is in the presence of another substrate it uses up that substrate first? I may be completely off with my reasoning here.
Even though it was weakly detected, doesn’t that still mean that the strain was able to used glucose? Or would you just ignore that data because it wasn’t substantial enough?
The other papers never mentioned a half-life, if I recall correctly. Does this make carbofuran significantly different or do all PAHs have similar half-lives?
Mineralization concerning PAHs hasn’t been discussed previously in the other papers. Does this make carbofuran significantly different as well?
To make sure I am understanding each step correctly- the process discussed in this paragraph was the initial step for the mutagenesis, correct?
Here is where the random mutagenesis process was completed and they moved on to RT-PCR, correct?
Being that the pink/light red color was more intense in the medium without additional nitrogen source, this basically means that when carbofuran is able to take up the nitrogen is changes in color similar to other PAHs turning blue/black?
“The selected mutants were tentatively assigned to five different groups” So basically, one genus but 5 different species of that genus?
Being that isolate sp. BAL3 was not conserved should it just be ruled out as insignificant?
If mutations affected protein expression of other genes does that mean the experiment needs to be start over?
What is the problem with categorizing individual disrupted genes by phenotypic affects?
The formation of carbofuran phenol begins the first step in degradation. Why would the mutants show no effects?
Is phenanthrene more or less easily degradable than naphthalene?
Is phenanthrene located in oil-contaminated soil? If we wanted to pull samples from the gulf like it was done for naphthalene could we do that?
I had this same question upon reading this section. Another possibility is a novel gain-of-function mutation, with analogous protein function to other extant PAH pathways in terrestrial bacteria. I also wonder if the ‘acquisition’ could be an activation of a pathway that may be just inactive in other marine bacteria? It could mean a lot of things, and makes me want to read the cited paper.
It seems like there are going to be a lot of results to read, as indicated by this statement. I am most interested to see a comparison of similarities and dissimilarities between the C. indicus PAH degradation pathway and those characterized terrestrial bacteria. Do they involve protein homologs? Do they produce the same intermediates and ultimate products? Are the products of the degradation waste products, or metabolites, or otherwise useful molecules for the organism? What is the energy investment into the degradation? Will the organism maintain the function of the pathway under stress? I hope to see the answers to some of these questions in the results.
Hi Annie,
From what we know about PAH degradation so far, I think it requires oxygen, because the enzyme RDH needs molecular O2 to break the aromaticity of one of the rings and start the process. There may be another enzyme that can do the same thing without O2 though, that would be interesting to learn about for the sake of comparison.
I don’t want this to count for one of my comments, but how cool would it be to get to go diving or in a submersible to collect sediment from the ocean floor for your project? That’s peak biology right there.
Does the term ‘metabolites’ refer to the products of PAH degradation only, or to intermediates in the process also? Is this technically an observational portion to the study since the strain they were growing and analyzing was all under the same conditions?
What is the goal of this knockout? I feel like I am missing some information – is P73 a protein involved in RHD? So the knockout mutant would be unable to metabolize PAHs? It seems like this would be a lethal mutation, for a bacterium that can no longer utilize its sole source of carbon and energy.
It definitely makes you wonder what kind of acquisition it was, of the three types we discussed in class.
So, in addition to the PAH degradation pathway, P73 can also metabolize many different sugars, but cannot initiate gluconeogenesis? I wonder what environmental factors would select for a bacteria with such a versatile ‘palate’ for carbon and energy sources?
A quick search revealed that Acetobacter aceti is a terrestrial organism, frequently sought out and used for its production of acetic acid from alcohol in fermentation of foods and wines. Makes me wonder how a gene that originated in this A. aceti ended up in sediment in the bottom of the Indian ocean!
This is the authors’ most interesting finding. A gene coding for a new protein that can initiate the ring hydroxylating dioxygenase step of the degradation pathway for toluene and biphenyl PAHs. As they state in the next paragraph, a marine organism that could degrade toluene would be a great tool for the cleanup of oil spills, since toluene is a toxic chemical that occurs naturally in crude oil.
BAD34447: large subunit of PAH-dioxygenase
CAG17576: ring-hydroxylating dioxygenase alpha subunit
ABW37061: Rieske domain; a cluster binding domain commonly found in Rieske non-heme iron oxygenase (RO) systems such as naphthalene and biphenyl dioxygenases
ABM11369: ring hydroxylating dioxygenase, alpha subunit
ABK27720: putative ring-hydroxylating dioxygenase large subunit
BAA21728: terminal dioxygenase component of carbazole 1,9a-dioxygenase
ABM11377: ring hydroxylating dioxygenase, alpha subunit
ABM11383: Rieske (2Fe-2S) domain protein
ABV68886: angular dioxygenase from fluorene-degrading Sphingomonas sp. strain
AF474963: Pseudomonas alcaligenes indole oxygenase-like gene, complete sequence; and putative dehydrogenase gene, partial cds
One question I could see the researchers pursuing in future studies is the control of the PAH degradation genes. Since P73 conceivably has the capacity to use multiple different carbon and energy sources, it would be logical it has some epigenetic capabilities to preferentially produce proteins to metabolize whatever substrate it was exposed to or growing on, right?
I understand that the mutants that didn’t produce a clear zone were chosen because it meant the mutagenesis process succeeded in mutating the gene. But why did they test it further with indigo if they already had what they wanted?
Why are they sequencing the plasmid at this point instead of transforming it into a host?
What factors helped determine that they should use a cosmid in stead of say a regular plasmid?
They concluded that the arhA gene was constitutively expressed because the the A4DR cells had low levels of arhA3 and arhA1 even when not induced. Could this be corrected by some of the methods we talked about in class like changing the inducer?
[These observations suggested that the genes involved in the initial oxygenation of acenaphthene did not function in strains AG2-45, AG2-48 and AG3-15.]
I get that a mutagenesis was performed and the strains were grown in different environments to determine whether they degrade acenaphthene. I don’t understand how the researchers cam to this conclusion.
I haven’t read about finding the regulator site in any of the papers we’ve read so far. How can knowing where the regulator binds be useful in further studies?
Could the OFRs that have no apparent connection to acenaphthene degradation have an unknown role in the cell? An experiment could be designed to knock out one to see if there is a change.
The scattered nature of the the sphingomonad’s genome seems to be an obstacle in identifying genes. What could be a possible explanation for its gene dispersal?
Does the mechanism for breaking down fluoranthene work for all the other types of similar PAHs? Wouldn’t they be different for every compound?
Is there a reason why high-molecular-weight PAHs aren’t studied as much?
How do they know if they have the right metabolite?
In the supplemental text, they found a lot of genes but I’m having a hard time relating them back to the initial goal of the experiment. Can you explain?
Here they are identifying the PAH ring-hydroxylating genes.
Oddly, these genes are in the same place. It hasn’t been this way with many of the other genes in the papers we read before. This is a guess, but I suppose different genus’s store their DNA differently than others.
Is there a reason for why their levels vary?
Does this mean new genes for the degradation of the PAH were found?
In comparison to the other studies, it seems like the information found in this part didn’t go as far as the other papers. In other papers this sort of information would be used to physically test if the conclusion was true or not.
Is computational genetics a common way of discovering the function of genes? Did the information used in the other papers rely on this kind of work to point the researchers in the right direction?
I want to know what separates these bacteria from other bacteria. What allows them to be able to degrade naphalene while others cannot.
This is a good point. The paper states that the pollutants are conversed to biomass, carbon dioxide and water. Which I guess either way you look at it, the remaining end products are less toxic or non toxic in comparison to the original pollutant.
I was thinking the same thing. Could these Naphthalene-degrading bacteria be used for clean-up of the BP oil spill in the Gulf?
Did they want to slow down the activity of their samples to make sure that the results they got were due strictly to experimental variables, and not the environmental changes while being transported?
Why did they choose to get sediment samples from 1-12 cm? Is this the depth where most of the oil had settled in the sediment? And why are the seawater samples taken from 15 cm? How big of a difference does it make in the results if these samples were chosen from different depths?
I was curious about this too. You would think they would be closer in range. Thank you for the clarification. It is true that we are all similar, but all so uniquely different.
I was interested in the GC-FID method, because I wasn’t sure what this was, so I researched it. I read that it was developed for analysis of oleic acid and fatty acids. What do these have in relation to nephthalene-degradation?
I am interested to in how the gram-negative can tolerate PAH better than gram-positive. I’m assuming it has to do with the peptidoglycan layers of the gram-negative.
Is the isolation of the naphthalene-degrading bacteria process similar to that which we performed in lab. Just test using different agars that they may possibly grow in until they get pure cultures?
What is the difference between substitutes and unsubstituted PAH’s?
Since this paper is saying that marine bacteria may be more significant in degrading PAH’s than terrestrial bacteria…. does this mean terrestrial degraders are limited only to land and vice versa, or are there some bacterial PAH degraders that are present in both marine and terrestrial environments?
The genomic library is used to represent all of the genomic DNA from an organism. This is done through DNA extraction in which the DNA is then cut to certain sizes. They are then inserted into vectors which are taken up by hosts. These hosts can then be used to amplify. The genomic library is used for sequencing genomes.
I looked up what agarose gel electrophoresis was, and I read that once a DNA molecule is digested, this method can then be used to see the fragments, because the electrical current moves the molecules across the agar.
Why are homologs for oxygenase subunits located in pairs usually? And what do you think the reasoning for the separation/absence is here in phnA1b?
A biotransformation is just a change or alteration, correct? What is the goal for biotransforming the substrates?
** My second comment is in supposed to be posted on paragraph 18 **
Today in lab, when you drew the picture and explained how the clones can be an “overlap” of a region really helped clarify the idea of having the same 10.5-kb Sau3AI fragment.
I think the significance of not finding the gene clusters for PAH degradation would be that the organisms lacking the gene cluster could not degrade PAH’s.
What is the importance of the major cluster? I get that it shows divergence of a-subunit sequences and similarity of amino acid sequences, but what exactly is meant by the major cluster?
I think that it would be better conservation to have one pair that can partner with many different dioxygenases.
I was researching this topic and read that arsenate is toxic because it can uncouple glycolysis, is this true?
I was wondering the same thing, especially since they included “surprisingly”. I mean they did have aerobic As(III) oxidation activity, so why would there be no authentic PCR products?
Why did they choose sites with differences in temperatures?
Aren’t cyanobateria a type of photosynthetic bacteria? If that is the case are only photosynthetic bacteria the only type of bacteria of interest when it comes to arsenic use?
I researched this after reading your question. DNA libraries are made from fragments of DNA. Where cloned libraries are made from cloned, reverse transcriptase of mRNA. So cloned libraries do not actually consist of DNA sequences.
Going through this figure today, it really helped see what was actually going on in the figure. While the Arsenite is in light it is being oxidized, then when it is in the dark, it is being reduced.
Even though we went over this figure in class, I still don’t understand why they used the three different variables of 100% H2, 2mM sulfide, and no added electron donors. What are each of these three variables offering us? I understand that they are driving forces for reduction, so is it just offering us information on which is the greater of the three for Arsenate reduction?
I was wondering the same thing, because here it doesn’t really help us to get a clear conclusion on specific microbes. But I guess that could allow for a follow up experiment using the techniques we will discuss on Tuesday.
In response to Rhyann’s comment… Is it because photoheterotrophy was responsible for the oxidation, and acetate was the electron donor for photosynthesis?
In my opinion this is the most interesting paper we have done this semester. I enjoy reading about the links made between our anoxic Earth vs. oxic. And it now makes sense that more recent biological communities would share relation to As(III) oxidation and As(V) reduction.
In reference to the deoxygenation and monooxygenation in this paragraph, is this what you were showing us an example of last Friday?
They can determine that the ring hydroxylating dioxygenase is the initiator by doing the same processes we are doing in class this semester?
So what is the point it “knocking out” the PAH degradation gene? It is just to make sure you are pin-pointing the correct genes involved in PAH degradation?
That sounds correct Daniel, because in the last sentence it says that the metabolites were determined using the GC-MS.
These are very extensive figures. How do you even begin reading maps like these? Or are they strictly read by computers?
I didn’t even initially think about genes being horizontally transferred to increase PAH degradation.
This is an important finding since it correlates with aromatic compound uptake.
The COG numbers ranged from 134-3. When it mentions poorly characterized, which is it referring to?
Is there normally a correlation between multiple antibiotic resistance regulators and PAH degradation pathways. I don’t remember reading this in our papers from last semester, but then I also wasn’t sure if I over read it.
They found 6 ring hydroxylating dioxygenase genes. Just because these code for this, does that mean they all actually assist in PAH degradation?
What kind of further studies could they do to study the functions and mechanisms of the P73T strain?
What is meta cleavage? and what is protocatechuate?
In PAH degraders, is it more common for the PAH catabolic genes to be arranged in discrete or dispersed throughout the genome? I’ve never really thought about which arrangement is more common/likely.
Is the topic of interest here, why PAH dioxygenases can oxidize fluorine, but not use it as a sole carbon source? Or will we discover that along with determining locations of PAH degrading genes?
Thank you for sharing this Molly. Background information is always a plus. Dr. Ni Chadhain, what is GSL and LPS? I remember hearing you mention LPS in class today or Monday, but I can’t remember what you were referring to.
The southern blotting made much more sense when you went over it today in class. I did not quite understand about wanting two different sticky ends while also containing the RHD probe, but you clarified this concept.
So, from what you were saying in class today, this part of the experiment is basically just to tell us yes you can force the E. coli to make the protein, or no you cannot?
Is the answer that cells use pyruvate during glycolysis to produce acetyl CoA?
This is what you were discussing with us in class today, that sphingomonas have some genes from gram positives.
What is meant by angular attack mean? It is pertaining to stereospecifity?
The southern blot technique made more sense after you explained it in class, as well as the “walking” done in designed primers.
Are there different types of dioxygenase genes? Like does each PAH degrader have different types of dioxygenase genes or are all dioxygenase genes the same because they do the same job?
So are you saying that even though the RHD is present, it doesn’t mean that it will utilize the PAH as an energy source? Does this mean that the degrader will use whatever route is most catabolically favorable?
When they mention the band extraction are they referring to agarose gel electrophoresis or southern hybridization?
The glass powder technique is pretty interesting.
How do you choose which strain of E.coli you want to perform a transformation with? Is it just based on availability or are there certain strains that work better than others?
Thank you for the information on the shotgun cloning Bentley. As I was reading that was my first question.
Homework 4 helped with understanding the information in Table 1. I understand what the table is actually talking about after performing our own BLASTs and listing our ORFs.
What is the importance of retention time in gas chromatography in comparison to expression of dioxygenase activity?
Can you better explain this sentence please? I’m not understanding how they can serve as substrates, but cannot grow on them. “Although strain A4 is not able to grow on naphthalene, phenanthrene, anthracene, and fluoranthene, these substrates could serve as substrates for the ring-hydroxylating dioxygenase system from this strain, since their corresponding cis-dihydrodiol products were detected in the resting cell reaction experiments.”
I’m surprised that monitoring PAH-dioxygenases in sphingomonads isn’t possible, because I feel like the species came up a lot in the BLASTs we did for homework 4, so it seems to be a well studied species.
I know that xenobiotic are something that is foreign to the body or environment. What is an example of a bodily xenobiotic? Are xenobiotics hard to degrade? I’m assuming degrading capabilities are based on what the foreign substance is.
When finding gene clusters of PAH degrading genes, how does this work if they are scattered out? Do they all have the same sequences, so when using certain primers, they all show up? Or does the DNA have to be cut multiple times in order to find every cluster throughout the genome?
I would think so, because if you find the genes involved first, you can then focus on the pathways they carryout.
I tried to look at the primers they used listed in Table 2, but it sent me to an online journal site, and I could not see the table because it only shows a preview of the article.
I think that transposon mutagenesis is really interesting, and it is probably my favorite part of what we have learned all semester. And I was really interested in our plasposon mutagenesis experiment in class, even though it did not work like we hoped.
Why did they have double crossover recombinants? And what exactly does that mean?
Why do some strains produce little indigo formations such as strain A4-PCM1(pBBadA13), and other like A4-PCM1(pBBadA14) produce it significantly? What causes this difference? are they metabolically utilizing other things available in the agar first?
I’m glad that were are getting to perform RT-PCR in our own class experiments. I read on the ThermoFisher website that RT-PCR is so powerful that it can “enable quantitation of RNA from a single cell”.
Why are they saying that the loose operon structure seems inefficient?
How can they find the genes responsible for further degradation of acenaphthene if they are dispersed throughout the genome? How do you even determine which step is next in the pathway?
This could be a stupid question, but it has me wondering. Since this is concerning oil in the Persian Gulf, it makes me curious after the oil spill in the Gulf of Mexico. Could these Naphthalene-degrading bacteria be found in in the Gulf of Mexico?
Why did the researchers choose this particular oil spill? Is it a special type of crude oil, or did the environment in which it is found play a role in the selection?
Why did the researchers decide not to add an IR Spectrum to the analytical chemistry? It would help further narrow down any isomers that could be found in the tested substance.
Is the purpose of this research to identify PAH degraders or is the purpose to explore PAH degraders and their effectiveness in bioremediation of the surrounding environment?
Could PAH degraders be under-researched because of the lack of equipment or resources available for such exploration? 2000m is a long way down and we definitely cant send a human down there.
What measures did they take to ensure that the extremely low temperature did not kill the bacteria that was isolated?
Later in the document E. Coli is used as a standard for unmarked DNA, does the first sample site act as the standard for the PAH degraders that are pursued in the second sample, or does this experiment assume PAH degraders are found dispersed throughout the sea floor?
Where does this Se(VI) and Se(IV) at high valence states come from? Is it waste from natural processes or is it pollution related?
Is there an aerobic component to E. cloacae? Is this bacteria an anaerobe due to high competition of resources found in the ocean?
I believe that a transcriptomic/proteomic approach would be useful, assuming that the researcher already knows the sequence that they will be looking for. On the off chance that their bacteria has a new sequence that has not been found, then the screen would surpass the gene and mis identify the bacterium.
With the fnr gene knocked out of E. Coli would this cause a completely anaerobic metabolism for E. Coli causing it to only use the Se as a source of carbon?
There is no further mention of the ogt gene in the rest of the results section so is it safe to assume that this gene played little to no role in the reduction of Se(VI)?
Based on this section E. Coli appears to naturally reduce Se(VI), but at a lower or less efficient rate than E. cloacae. Would this be the reason they did not try to incorporate the fnr gene from E. cloacae to E. Coli to prove that the fnr gene is responsible for the reduction of Se(VI)?
For E. cloacae to have a real world use in the reduction of Se(VI) that could reach toxic levels in an aquatic community, would there need to be a mutation of the fnr gene where Se(VI) is reduced in the presence and absence of O2 or would the rate of reduction be significant enough with out it?
Would these proteins be a way to make E. cloacae a suitable candidate for real world applications where a Se(VI) reduction capable bacteria would be needed for biosynthesis? Could these proteins be transplanted into the genome horizontally or would they have to be transplanted virally?
I am interested in the amount of arsenic V that was reduced to arsenic III.
Wouldn’t this be a good opportunity for the authors to employ transcriptomics? What would be the benefits of employing the experimentation in the lab versus out in the field.
It is quite amazing how versatile rhodococci is and how it is able to degrade environmental pollutants that are too difficult to degrade for other organisms. While the rhodococci can metabolize xenobiotic, and halogenated compounds, which can be found in various herbicides. Is it possible for the rhodococci to metabolize other sorts of controlled substances such as pesticides or possibly fertilizers?
It is very interesting to learn that during degradation Rhodococcus uses aromatic rings that catalyze the initial reaction of bacterial degradation. During this degradation it uses two subunits, one of which is the alpha subunit that aids in electron transfer to the dioxygen molecule. But the other subunit, the small beta, was not explained further. Does this subunit have a purpose similar to the alpha unit? Or does it perform it’s own necessary task in the degradation process?
I think it is really interesting that the sample chosen was from an oil-contaminated sample in Japan. The fact that the oil could impact the growth of the culture compared to if the soil from that area was not contaminated. It makes me wonder what kind of oil was present in the soil and if it was contaminated with a different kind how would the affects differ.
I believe that OD600 is an abbreviation for absorbance or more specifically optical density. In this instance the sample was measured at a wavelength of 600 nm. This method helps in estimating the concentration of bacterial cells in the liquid.
I think it is really cool to be able to visualize the relation between the different mycobacterium and where Rhodococcus falls in the phylogenetic tree. It makes me wonder how other closely related would react under the same experiment or even their common ancestor.
I was thinking the same thing Aubrianna! It’s really cool to think that I am able to understand the paper better after every lecture and lab day. A lot of these terms and processes I had no idea what they were about but now I can formulate a general idea of what is happening during this experiment.
Since the Rhodococcus only exhibited an amplifies 100-bp band compared to its similar counterparts exhibiting a 78 bp band, would it not be expected that the Rhodococcus to perform the way it did? What is so different between these genes that allows Rhodococcus to do what it does?
It’s really amazing that a new naphthalene dioxygenase was reported in Rhodococcus during this experiment. Since it only shows a distant relation to the pseudomonas enzyme I hope to read more papers about this dioxygenase and its capabilities and be able to see what it is capable of in other situations.
It is really interesting that these PAH’s can be brought up the food chain and affect humans. I wonder what kind of effects they have on humans and what some of the symptoms would be to in order to diagnose a patient with being affected by PAH’s.
While it suggests that obligate marine bacteria may have more significant PaH degraders, could the physical parameters of their ecosystem be helping them? Terrestrial organisms have a restricted form of movement through their ecosystems where substances can only be moved via wind, water, or other organisms, but in marine ecosystems the movement of substances is much faster and less restricted possibly exposing the bacteria to more PAH’s.
I understand that the Marmur method allows DNA to be isolated from microorganisms and keeps the needed material active, and polymerized while keeping the majority of proteins and RNA away, but is there anything else about this method that makes it a necessity for this experiment?
So in this case, the important thing to remember is the fact that the converted products were being identified and measured as well as their purification that was produced in the culture supernatant, correct?
In the flanking region of phnA1b it is said that there is no present small-subunit gene in the oxygenase pair. Was this subunit lost in transformation? Also how does this hinder the genes functionality throughout the cell?
Tara, I also looked up P. stutzeri and found out about the spinal fluid and it’s similarity to the NADH ferredoxin. It’s really interesting that they are similar and it would be cool to be able to learn more about the history of the relationship between the two, just like you said.
I also have the same question Jesse. Is there anyway to access the Figures they are referring to in the text? I believe that if I was able of view them I might have a better understanding of what they are trying to say about their trees and how that relates to them falling out of the cluster.
Jennifer, I think it would make it more efficient. Since the PhnC is involved in both aerobic and anaerobic pathways, not only is it degrading the compounds fast but it is also able to degrade them when there is no oxygen is available.
Hey Alexandria, there are a bunch of other relationships between cyanobacteria and chemolithotrophs, and even some pretty large eukaryotes too. In some cases the cyanobacteria will provide its host with fixed nitrogen and the roles can even be switched around where the cyanobacteria is being provided for. So it sldo makes me think the same thing about whether or not they might be breaking down the arsenic to help each other out.
I was also thinking the same thing Louie. I can’t wait to continue reading this paper, it seems like a lot of the information will tie into some information from the Biology of Algae class that I took and how cyanobacteria could have nitrogen fixing activity during daylight hours if induced by low ammonium. I wonder if that sort of reaction will be shown in this different type of environment.
I agree Tim, identifying the protein produced would definitely help in figuring out which genes are responsible for the arsenic cycling or resistance quality since some of the PCR primers were not giving us enough information.
Caleb, I was also thinking about the environmental parameters of these anaerobics. Since the temperature range is so broad, it makes me wonder if they hold any sort of extremophile qualities, where their tolerance can withstand very cold or hot temperatures. It’ll be interesting to see the phylogeny tree and find out if they are related to any temperature tolerant organisms.
I wonder what kind of pigments these arsenic cyclers have, considering that they undergo anoxygenic photosynthesis it can be assumed that they use bacteriochlorophyll, but determining which kind might help with understanding the limits of their photosynthesis.
It would be interesting, considering the technology available now, if they redid the experiment with a omics approach to annotate the gene that oxidizes AS(III).
I was confused when I first read this paper as well. Thank you for your definition! This concept seems interesting, and I appreciated the explanations in the text to both co-metabolism and the use of consortium strains. These definitions helped me understand the biological aspects of bioremediation as it relates to microbiology and how it can be efficiently used to preserve the environment. It shows that there are natural ways that we can effectively target toxic oil spills instead of introducing harsh methods that could potentially devastate the environments even more.
I wondered the same thing, especially since there has been no further study for PAH degrading bacterial strains from the Nakheel region. Were there other factors contributing to the unique degradation patterns besides the low nutrients and high hydrocarbon pollution?
It does not specify that a control was collected. Is that already assumed, or is there even a need for a control in this type of experiment?
The paper does not specify that a control was collected. Is that to be assumed, or is there even a need for a control in this type of experiment?
Why would conductivity of the sediment samples affect the PAH-degraders? Do conductivity and pH affect each other, or were they measured together for the sake of efficiency?
Does it matter how much PAH they end up with? It seems as if they would not have a large number of PAH left after this rigorous isolation process if they began with 1% PAH. It could be assumed that it would be a big inconvenience for them to repeat that process if they did not end up with a desired amount of PAH.
Why did they construct the phylogenic trees? Was this to organize and assess the complete variation of the bacteria present? Or did this help them in some way analyze which bacteria would degrade the toxic oils more efficiently?
What would the difference between the gram-negative and gram-positive bacteria have on the effectiveness of the PAH degradation? Why would that make a difference?
I was wondering the same thing. It would seem that LB would be more useful, but that conclusion cannot always be applied in such seemingly obvious situations.
Was this step in the experiment the reason why the phylogenic tree created earlier on in the methods portion of the paper? Does this make identifying effective bacteria and related bacteria easier? It seems that this could help find possibly both cheaper and mire effective cousin bacteria.
I too thought of your first question, if related bacteria are just as effective at PAH degradation as their cousins.
Since P. aeruginosa is possibly unsafe to test due to human pathogenic qualities, would there be any instance where that could be tested in a safe environment using technology? It begs the question of if it would be ethical or worth exploring other testing methods if it means finding the most effective PAH degrading solution, though it would be ethically irresponsible.
I had the same question, Cody. Does this mean that the other isolates can be defined as the bacteria that they are so similar to, or are the 1% and 2% differences significant enough for them to be labeled as something else?
It’s interesting to note the drastic difference of the 3 isolates on degrading pyrene vs phenanthrene. They all performed similarly on pyrene, but there are varying levels of effectiveness on the phenanthrene. In both cases, most of the isolates seem to follow the general pattern of the control and follow its fluctuations, just at a much higher degradation % rate.
That was also my first thought upon reading this section. After learning what turbidity and optical density are/consist of, it also made more sense that the solutions would be clearer if there were larger microbes present.
What is the purpose of the bacterial mat? It is mentioned here and in the next paragraph. Does it influence the temperature of the microbes? And would the absence of the microbial mats allow for more effective extreme cooling as apposed to them being present?
What is the purpose of using the different carbon isotopes 12, 13, and 14? I see that all 3 are used throughout the experiment. Do they relate to the bacteria being degraded, or do they relate to the methodology to extract the strains?
I agree, Kelsey. It seems as if the researchers obtained different results then they originally hypothesized. Once they identified the most efficient mineralization technique, they were able to continue experimentation to see which compounds would be degraded best by which means. It is clear in the graph in paragraph 2 that the 4567-24 has a much higher percentage of initial CO2 concentration captured.
It seems as if there are drastic differences in both the 14C-labeled PHE and the HPLC regarding the live cultures vs the acid-inhibited controls. Their effects are opposite of one another.
The PAH performed better with live cultures, but poorly in the control group.
The HPLC performed better in the control group and more poorly in the live cultures.
Was this effect to be expected since the PAH was removed in the HPLC?
How would the oxygen levels in the cell in the medium show selenate reduction? Would the less oxygen show more reduction?
What would the importance of determining the shape and size of the selenium particles formed be? Is this to determine how much of the selenium is degraded?
I had the same question. There didn’t seem to be a clear reason as to why one cosmic clone was chosen over the other or that any evidence of that would have been indicated later in the next paragraph or the rest of the paper.
How was the fragment pEC223 chosen from the 4 sub-clones? Does that have any significance as to which genes were isolated for selenate reduction?
If the FNR gene operates best in anaerobic conditions, how would one be able to monitor this in a natural environment?
Was the strain K-12 introduced to show that there are other means that E.coli can use to reduce selenate without the presence of the FNR gene?
Is this paragraph saying that the FNR gene was needed for the S17-1 strain and the YgfK,M,N gene was needed for the K-12 strain?
I would presume that the hydrocarbon degraders would degrade the oil differently, even if the oil in the surface waters is the same that is in the deep sea. The extreme environment of the deep sea would cause these bacteria to rely on other energy sources, possibly chemicals such as sulfur, etc. How much of a difference would this make on the effects of the hydrocarbon degraders?
I also made that observation, the extreme living conditions cause me to believe that these deep sea bacteria are a different strand than the coastal bacteria.
How would the phylogenetic identity of am organism determine its function, in this scenario with PAH degrading bacteria? I understand the concept of creating a phylogenetic tree for the purpose of identifying related and efficient PAH degrading bacteria, as conducted in the last paper. How would one determine the function of a certain phenotype or genotype of a PAH degrading bacteria?
How different are the microbial reduction of the selenate and selenite vs the abiotic reduction? Are they equally effective, or would one be much more effective than the other?
Would the bacterium being gram-negative be significant in its metabolic functions, or is that just a way to further identify the bacteria?
Does the process of Degradation depends upon Number and kind of micro organisms or the Chemical property of the PAH which needs to be degraded or both?
As Microbial degradation of PAH is quite interesting topic, I also read few other short articles about this, and I would like to share some information here. Prolonged exposure to chemical toxicants can cause adaptations in microbial populations that results in greater resistance to toxicity or enhanced ability to utilize toxicants as substrates for metabolism ( source of energy). Actual mechanisms for these adaptations are still unknown.
Does this biodegradation takes place in the absence of Oxygen also? I mean in anaerobic conditions ?
I am excited to learn more about PAH degrading bacteria and bioremediation. I hope many of my questions will be answered in the upcoming sections.
I wonder what if gene insertion(from other PAH degrading bacteria) is done, will be the affect of degradation of PAH increase or decrease or remains same!.
Does C . baekdonensis B30 strain degrade high molecular weight PAHs?
In this context , I think there are referring to the intermediate metabolites in fluoranthene degradation only.
I just want to know how the mutant work, what will be the results? I hope I will find my answers in coming sections.
As far as I remember it is around 1%-2%.
Why is there a differentiation in 48tRNA gene represent 19 amino acids and a tRNA for other amino acid? why not they considered 49tRNA and 20 amino acids?
Does COGs transfer between the generations or are they differ among the generations of same species?
I was thinking the same! I’d like to find out if there was potentially any type of symbiosis between the two!
I feel like the dark reductions occurred without a light source, which might cause it to have fluctuating temperatures as the experiment wore on from day to day. However, in the light controlled experiments, I think that the ability to control the temperature was easier as light was a component of the experiment!
I think the same! I wonder if the microbial populations were dependent vs. independent on light and if mutations had occurred in order to select for light oxidation and dark reduction.
It is definitely cool to see that hydrogen sulfide stimulated both the anoxygenic photosynthesis and in the dark incorporation of acetate. I think this makes sense because both reactions use sulfide as the beginning electron inorganic donor.
Since the bacterium rhodococci are capable of biodegradation of environmental pollutants, I wonder if it is capable of a mutualistic relationship with other organisms? Its ability to metabolize xenobiotic compounds that are not naturally occurring is quite unique and poses the question of whether it can exist symbiotically in different environments.
I was wondering the same? What is the specific function of the small beta unit? To my understanding, the alpha unit is involved in electron transfer at the iron catalytic domain. Does the beta unit function in reduction as well?
It is very interesting to note that the two ringed and three ringed PAHs are more readily degraded than the four or five ringed PAHs. I wonder if this has anything to do with the chemistry of the PAHs. Also, how can the enrichment be better modified to target four and five ringed PAHs?
I definitely noticed that too. I wonder if there was not continual enrichment and subculturing of the Rhodococcus on Fla would have resulted in different outcomes? What if the cells were instead subcultured with Nap or Phe?
It is very interesting that the attempt to enhance Nap, Phe, and Fla degradation actually resulted in their inhibition or had insignificant results. This can prove to be a beneficial discovery for producing a napthalene-inhibiting substance.
Since there was no homology between the tested rhodococci ARHDs in this study to any prior studies, does this quantify a new type of dioxygenases unique to this species of rhodococci? Are there any additional tests to confirm this?
I agree with this. It’s very encouraging to see education in real-world application. This studies shows that there are methods to inhibit degradation of certain compounds like Nap and methods to increase the biodegradation of Fla.
PAHs seem to be a by-product of factory and industry activities. Is there a way to regulate these processes or regulate the release of PAHs before it enters the natural environment?
Since Cycloclasticus operates in a marine environment, this bacterium must be salt-tolerant. Although I had no growth on the mannitol salt agar plate in lab 5, I wonder if I isolated an NP-degrader from a water sample if I would have growth on the mannitol agar and if it could possibly be a strain of Cycloclasticus?
Can PAHs also be transferred to humans through respiration or contact? Does it have to be ingested?
Were the dioxygenase proteins tested for the presence of the substrates or were its activity actually measured? How was it measured and quantified?
I was wondering the same! Perhaps it may not have an effect at all? Maybe the presence or absence of this gene just serves as a comparison point for the two? I wish there was more information about the small subunit!
What does the data mean by “agreed well”? Does this conclude that, yes, the phnA1A2A3A4 genes actually did code for the holoenzyme of the aromatic dioxygenase? Is there an explanation for why the data may be a little off from the predicted sizes of Phn?
I think so too! Is there a common ancestor that we can compare some of this data too since the basic sequence of features are conserved?
Since these proteins may be multipurpose electron transport proteins, would there be high regulation of these proteins since its functionality is enhanced?
I’m wondering if there is a specific reason as to why the activities of the fused, unfused, and sing-ring compounds were not different in these situations. Does it have to do with hydrogen bonding?
I’m wondering if these bacteria can use arsenate and arsenite as an electron donor in major pathways like glycolysis or the electron transport chain due to its high potential?
From the last sentence of this paragraph, are they suggesting that there are certain bacteria that can undergo “arsenification” and “dearsenification”? Also, what would be the ecological importance of forming arsenate and arsenite if they are toxic compounds?
Okay, so reading further, I do indeed see the the suggested coupling of arsenic metabolism. I wonder if one part of the couple is to recycle arsenic under anoxic conditions, will this also be tested under oxic conditions? Also, will the recycled products from this arsenic metabolism still have toxic effects on the environment or will they be neutralized?
I think that this is very cool that they took samples from rocks and cobbles that we would see on everyday hikes. This just reminds us of how much microbes rule the world! But on a more serious note, why was the slurry stored in the dark? Were the biofilms found in shady/dark areas?
I wonder why there is a distinction between the light incubated and dark incubated bacteria samples. Do they exhibit phototropism?
I was wondering the same! Does it have something to do with the photosystems and the light dependent and light independent reactions?
I feel like they mean that out of all the clones they did, there was only one clone that matched the arsenic oxyanion translocation pump gene.
I, too, agree that it is interesting that the sample was collected from a city in Japan. Why was Japan the site of sample collection? I wonder if there would be a difference if the sample was collected from an oil field in the Middle East. What type of soil and oil is optimal for the cultivation of PAH degraders?
I wonder why gas chromatograph was used as the method of analyzation. Do the bacterial strains give off a certain molecule as it degrades the aromatic hydrocarbons? Could a spectrophotometer also been used as a qualitative measure of analysis for bacterial growth?
In paragraph 9 of the introduction, it was mentioned that present study and isolation of Rhodococcus does not have homology with catalytic domain of any prior ARHD Rhodococcus species. In this study, it is reported that the isolated bacteria exhibited 99% homology with the Rhodococcus species in the 16S rRNA gene, which is a highly conserved gene region that is used to identify microbes in complex environments. I wonder, if this bacterial strain exhibited any homology with the Rieske center of ARHD’s as well. Does it also have homology with the catalytic domains of prior ARHD’s of Rhodococcus species?
It appears that Fla degradation followed a logistic rate according figure 2. I wonder how the degradation rates of Nap and Phe compares to Fla on a regression curve. I wonder what the difference in molecular structure led to the high rate of degradation of Fla in comparison to Nap and Phe. Knowing this, is there any way to select for the degradation of Nap and Phe better?
Is there a particular reason why the ferredoxin and ferredoxin reductase genes were amplified through PCR rather than another genetic set? Is this code unique to the bacteria?
I wonder how the results would turn out if PAH samples like from the previous paper and from our lab experiments were used instead. How would the collection of a variety of PAH samples have an effect on the results?
Is the degradation process of high-molecular-weight PAHs unclear because there is a lack of research in that aspect, or have researchers tried to study it but were unsuccessful?
If the P73T strain is the first fluoranthene-degrading bacterium to be found within the Rhodobacteraceae family, would that suggest that there may be other bacteria in the family that could degrade PAHs that we haven’t discovered or haven’t studied enough yet? Also, could the PAH-degrading genes be moved from P73T to other bacteria within the family using horizontal gene transfer?
What makes fluoranthene different from other high-molecular-weight PAHs that it is a good model? The previous paragraph mentioned that the degradation of high-molecular-weight PAHs wasn’t fully understood, so I’m curious as to why fluoranthene is the model of choice. I’m also intrigued as to why fewer fluoranthene degraders have been isolated from the marine environment. Is it because there is a lower fluoranthene concentration in those environments, or because bacteria marine environments aren’t the preferred research specimens?
I’m also wondering about this. Especially since the degradation of the high-molecular-weight PAHs is not fully understood. Some bacteria can degrade high-molecular-weight PAHs and some can, so is it the PAHs themselves that cause this, the bacteria doing the degrading, or both?
Me too. The fact that only a small percentage (like 10% I’m pretty sure) of the Earth’s oceans have been mapped and researched always has me curious about what else will be found in the ocean as time goes on. There’s so much to be found and researched from and about the oceans.
The mutant strain was generated to analyze and compare to the wild strain during experimentation. I’m curious to see if they will only try to examine the differences between the mutated strain and the wild strain, or if they will be trying to amplify the PAH-degrading genes in both the mutant and wild strains to see how the mutation will affect the function of the gene.
If fluoranthene is a PAH and is the only source of carbon and energy for the bacteria, then the mutated strain should starve and die since it is unable to degrade the fluoranthene. I’d like to see if this will be the case, or if the mutated strain will have an alternative PAH-degrading gene that will allow it to survive.
The coding density of the human genome is about 3% from what I know.
I thought it was really interesting that so many horizontally transferred genes were found. The P73T strain using HGT to adapt its PAH degradation is not something I expected, as I didn’t ever consider bacteria able to quickly adapt like that.
This answers a previous question of mine. I was curious to know if the bacteria with the mutated gene would be unable to use the fluoranthene, or if they would have an alternate PAH-degradation gene that would be used.
This is the same thing I was wondering! I looked up a few things, but there are mixed answers from what I have seen so far. I would like to know a for sure answer as well.
What makes PAHs become resistant to biodegradation?
So if non-sporulating aerobic rhodococci are commonly found as biodegrading environmental than could they be used more prevalently in the world to break down manmade pollutants?
I find that it is interesting how different types of bacteria take different types of stains. It is important that we can use stains to distinguish different types of bacteria.
I wonder how much Fla would have been degraded if the samples had been allowed to incubate for longer amounts of time. For instance, how long would it have taken for Fla to completely degrade.
I wonder why the Rhodococcus sp. was able to degrade only that much of the ILCO in two weeks. How long would it take for the Rhodococcus sp. to degrade more of the ILCO.
I find it interesting how the writer chose to focus on how PAHs are present in the marine environment instead of the terrestrial environment. Perhaps this paper will focus more on how PAHs affect marine organisms.
I would be interested in learning more about how this genus of bacteria, Cycloclasticus, can degrade petroleum PAHs. That seems like it could have important real-world applications if used in the right way.
If less pure chemicals had been used I wonder what the consequences would have been to the results.
I wonder why the Luria-Bertani agar plates were chosen for the plasmids and to why there were added antibiotics. Also, why was that concentration of antibiotics chosen for the plasmids?
I may have missed this but what is the flanking region?
It is interesting that the researchers were able to show that PhnC is involved in the upper and lower pathways of degrading naphthalene, phenanthrene, and biphenyl. Since it’s involved in the upper and lower degrading pathways does it mean that these are more efficient in degrading these molecules?
It’s cool that the PhnC is able to use both the upper and lower pathways to degrade naphthalene, phenanthrene, and biphenyl. Since PhnC uses both pathways does that make it more efficient in degrading naphthalene?
I like that this paper is different from the other two papers that we have done. It is cool that there are bacteria that are able to use arsenic to survive when it is so toxic to a lot of other organisms. It is even more interesting that the bacteria are able to use the Arsenate and Arsenite to create energy for themselves.
I am also interested in learning why their PCR failed. Perhaps it was because the primers were not right for the genome. I wonder how it would be different if the researchers had taken samples from a different biofilm that had similar bacteria. I am interested in learning about how these different bacteria work to get their energy since it is an anaerobic system that is light-driven.
I wonder if the results would change if they took the bacteria samples from a different hot spring with the same conditions?
Maybe the slurry did not need to be intact because it was going to be used for growing the biofilm on media so they needed it in a liquid state. But that just a guess.
I think it was interesting that there were only two main types of bacteria that were found in these biofilms. It makes sense that several types of Archea were found in the biofilm since the biofilm was taken from a harsher environment.
I found it interesting how the As(V) and As (III) had nearly opposite reactions when introduced to the light and dark regimens. I don’t think I was expecting that to happen.
It is interesting that Rhodococcus has the potential for such varied uses. I would be interested in learning about what kinds of applications it has in environmental remediation.
I agree with you that Rhodococcus could have great benefits for the environment. It would be great for the many uses of Rhodococcus to be recognized and well developed in the coming years.
I also found it interesting how the addition of the YE resulted in the inhibition of Nap while did not really decrease the degradation of Phe and Fla.
Why did the YE initiate the early degradation of Fla? Looking at the rest of the paragraph it certainly sped up the degradation of Fla during the incubation period significantly.
It is interesting how the increase of Fla concentrations could be inversely proportional to is degradation as the paragraph does not make it very clear. But reading the other comments does help to clear it up a bit with how Fla could be toxic in high doses.
I find it quite interesting that Rhodococcus what able to degrade aromatic fraction of Arabian light crude oil and the aromatics and alkanes of motor oil. But in the present study why was only the aliphatic fraction of ILCO able to be degraded by the Rhodococcus?
I also find it interesting that a new naphthalene dioxygenase was found in Rhodococcus sp. NCIMB12038
It is interesting how a new naphthalene dioxygenase was discovered during this experiment in the Rhodococcus sp. This is cool as it only showed a distant sequence relationship with the Pseudomonas enzyme.
I am curious to see how the Rhodococcus could be used in the future for biodegradation of different hydrocarbons. This is a very interesting product but I believe needs further research to help develop the methods to effectively use it as a biodegradation tool.
I am also curious as to the age of the paper and what kind of implications that this paper has had and/or will have to the study of Rhodococcus.
Are there be any benefit to not converting some of the molecules to their cis-dihydrodiol forms or to having the other molecules stay in their monohydroxylated forms? From the reading it says that the second set of molecules mentioned stayed in their monohydroxylated form due to structural instability in the cis-dihyfrodoils. But could there be any other reasons for keeping or changing the molecules one way or the other?
How many of these kinds of degraders where found to have the ability to oxidize indole to produce indigo? Do the ones in our experiments use that since the colonies will appear blue.
I would like to know what makes filter mating better for this study rather than other mating protocols.
I was wondering the same thing as Hampton. Is this like a standard dilution or is it based off what they need for the experiment.
I was wondering the same thing while reading this. Is this a standard dilatation or does it go by the amount of cDNA solution?
Why would they not show the data for the Southern Hybridization? I feel like it that confirm the loss of the region that you would want to show that data?
This is because of the size. Plasmids are only approximately 10kb, while cosmids are for approximately 40kb. Now they could have opted for a Phage instead possibly because the phages are used for the same size, but plasmids are going to be too small.
So since they are studying a new strain here, I am assuming that they will be incorporating the techniques in the previous papers here to identify the genes and pathways?
Is there a reason why there is not much known on marine-sourced degraders? Are they harder to study or just not as common in studies?
So GIs are the areas involved in horizontal gene transfer correct? Would they not run into problems with this later if they are only studying these areas?
This is because they have the genomic islands which give them ideas of where things are located, if I am understand the method correctly.
I find it interesting that it was this was found to be the only pathway, in comparison to those who have two or more.
So for studying the function of the catabolic gene, would they just move on to mutagenesis?
Is bioremediation a process of introducing foreign microbes to the environment to breakdown the pollution? Is this considered a natural technique?
Why would they not isolate and analyze the bacteria from that location? Was it because of the lack of enough nutrients and the excessive amount of pollution?
Was the purpose of taking the temperature of the water to compare it with the abundance of bacteria found at each location?
Are the nutrients’ analyses vital to the decision of the bioremediation microbes selected to attempt to naturally correct the issues? Is that how biologists know from the get go if bioremediation is an option?
Why is the potassium in location B & C so high compared to location A?
I think gram-negative bacteria is only pink when performing “Gram Stain” tests, which stain the gram-negative bacteria pinkish red.
It’s interesting that the rod bacteria are favored over coccoid bacteria. I researched on the internet and a website said that the large surface area’s don’t necessarily mean a large adhesive force.
I believe that heme is a co-factor (prosthetic) that is required in hemoglobin and myoglobin for the proteins to function.
I believe the purpose of the study was to identify any other possible genes that could reduce selenium.
Knowing the morphology of the particles would tell us how the bacteria reacted to the transformation?
The rate of selenate reduction was measured because of the direct relation to the changes of concentrations, correct?
Did the colonies turn red because they were unable to reduce nitrate?
So, the exterior cell membrane of the bacteria would determine the morphology of the Se particles?
My takeaway is that information from each experiment helped them come to that conclusion after their own study results.
It appears that they are saying that FNR regulation isn’t necessary if certain proteins are present. So yes, I think they are saying there are possibly multiple ways to reduce Se(VI).
I was wondering this same thing. If we are exposed to PAH’s through digestion do you think that the way certain foods are processed could increase the harmful effects? Also, regardless of whether the food we are digesting is processed or not, it could still have been contaminated from the environment. While it is important in protecting workers who could potentially be exposed to PAH’s via respiratory uptake, I feel it is just as important for people to be informed that these are in foods we are ingesting.
I also find it interesting that these can be identified using the same techniques that we are going to have the opportunity to use in the lab. If we were to do this testing in an area with greater exposure to PAH’s do you think it would correlate with the rates of cancer in that particular area? This is something that I was interested in reading a little further into. This lead me to asking myself if these areas that have greater exposure to PAH’s are well informed of this and are aware of the effects they can have on the human body.
Is there a chance that a person can be contaminated with PAHs such as ingestion without you realizing it? I’m wondering if the effects are immediate or are they something that can sometimes be over looked, such as shortness of breath and dry cough. It is hard for me to believe that I have never heard of PAHs yet they are potentially everywhere around us and even in what we eat or drink. I see that it says prolonged exposure causes more serious side effects but once you are exposed to these do they stay in our bodies or does our body eliminate them?
I was also thinking this same thing. In our lab I’m interested to see what the difference in growth is between our diluted cultures and our more concentrated cultures. Along with this I am wondering the difference in R2A plates and MSM plates. Will they differ in growth, color, etc ?
I was also thinking about the amount of PAHs in the environment. It it alarming that Dr. Ni Chadhain found this just simply in her neighbors yard. Is the public aware of things like this? I feel that there should be more awareness of the harmful effects PAH’s have and also more awareness of their abundance in the environment.
After reviewing the results I was wondering the same thing. I feel as though this is a question we will be able to answer after completing this course. After finding out what differs in their molecular structure I feel that we would be able to identify a better way to detect for the degradation and not only these but others as well.
It is interesting that I can immediately relate this back to what we are doing in our lab. Although, we are only a few weeks in to our course I already can identify what the difference is in growing a medium in an MSM plate vs an R2A plate. Even before we began this weeks lab we were told to look for a blue/black colony on our plates.
I did not initially expect the results with the added YE to completely inhibit the degradation of Nap. This makes me wonder if there are any studies about different strains that could possibly be tested on Nap. While this study was successful in regards to the degradation of Fla, I feel that there should be more experiments done to see what strains could be successful with degrading things like ILCO. Obviously there is going to be a lot more to the composition of something like ILCO, but I think it would be worth the extra work to help protect our environment.
I am curious what exactly (with the addition of YE) caused it to be so successful in degrading Fla, yet completely inhibiting the degradation of Nap. The Fla degrader seems to be more complex as they stated that another study found there to be two clusters depending on their PAH utilization capabilities. Do we know which cluster the one used in this experiment belonged to? Which also makes me wonder would a strain from the other cluster have been as successful in this experiment?
I guess I expected there to be a direct relationship between the concentration and the amount of degradation. Also, in this section of the paper it doesn’t state what the time frame was for the 750 and 1000 mg1-1 degradation but assuming that it would also be 8 days, would a longer time frame allow this to have a more successful rate? Or does it reach a type of thresh hold
I am interested to compare and contrast between these and the Rhodococcus that we read about in paper 1. The first question that comes to mind is wondering if one of these are generally more affective than the other? I am also interested to see the difference in the results of their degrading ability on different PAHs. If these are used in costal marine environments is there a chance that they have been used around our area for any type of bioremediation?
If marine bacteria is potentially more successful at PAH degradation this makes me think that Cycloclasticus will be the more successful one when comparing the degrades from this paper and the last paper. Also, it seems that the success of their degradation might be effected by what type of environment they are in, and not only which strain etc. Would we be able to conclude that one environment supports PAH degraders better than another?
This is the overall question I have had since the beginning of reading these papers. In just a short time we have learned the multiple routes and the extent to which PAHs can effect humans, animals and the environment… yet before this course I had never heard of “PAHs”. This raises the question of how many people are aware of the extent to which we are exposed to them and also the extent to which they effect everything in the environment surrounding us.
I was also wondering this. I think it is realistic to assume that depending on the situation one may be more effective than the other. But when they describe either one as effective do the byproducts play a role? Furthermore, does one give off more toxic by products than the other? I feel as though this is definitely something that they would consider when using either Fla or Cycloclasticus in bioremediation.
Will we get to learn more about the specific endonuclease that they are using to do this experiment? This also makes me wonder over what range can endonucleases be used or if they have to be very specific to the DNA sections that they are cleaving. Also, what was significant about the size range of the fragments that they recovered?
When I think of centrifuging I always think of chemistry 131 lab. I never expected further education to reveal the importance of this and that we can actually separate cells by simply centrifuging. After reviewing our pre-lab it’s so cool that we are going to get to do this Wednesday! The idea of us digesting the RNA and being left with DNA from our own culture is exciting and it’s also interesting to relate what we our doing in our lab directly to these papers.
I was also wondering the same thing after reading this paragraph! I do feel as though it is saying that there would be a specific DNA sequence that gives microorganisms the ability to degrade PAH, but I could be misinterpreting the wording. So does the order of the genes have any effect on their ability to degrade PAHs? Or does it matter that the order of the genes were different from the previously reported genes?
I was also thinking this! I’m wondering what the effects of the opposite direction of ORF7 is in the homology since later in the paper it states that the order of the genes (that exhibited high degrees of similarity with the polypeptide sequence) is different than the order from that previously reported.
Since they are located on separate transcriptional units does that completely determine that the electron transport proteins are shared with other redox systems or is it just assumed that they are shared as the reason for why they are on separate units?
When I was reading this paragraph I also was wondering about what degree of significance evolution would have on this expression mechanisms. Since it is so highly organized I feel that this is something that would be very interesting to read on from an evolutionary perspective. Could scientist alter the gene expressions to see what increases and also decreases the expression of the specific genes we see here?
I am interested to know if the conditions of Antarctica and its wildlife have changed in the past year, with less travel/tourism and possibly less exploitation of natural resources that leads to spills; and if any change has been for the better for the organisms that inhabit it.
Why and How is Diesel oil the most commonly used fuel in Antarctica? I have always known the continent to be a “cold and snowy desert” with nothing and no one around.
I wonder why they chose to only take 4 samples from each site (exposed and not exposed to diesel fuel). I would think they would want to collect more samples from the sites to have a larger sample size.
Why did they choose 168 h to grow the cultures? and Why was E. coli used as a control in this experiment?
I find it interesting that S. xenophagum did not show a chemotactic response toward any stimuli used. Is this due to it not having the proper mechanisms? Does this microbe have chemoreceptors that can detect cues? Does it have flagellum that could help it move toward a cue? I tried to Google this strain to see what the cells looked like but was unlucky.
Wow! I think it is very cool that Salicylic Acid is produced in this process. This is a chemical that I am very familiar with in my day to day life (in skincare) and I know that many people are familiar with it’s close relative- Acetylsalicylic Acid aka Aspirin.
Can these bacterial isolates work outside of extreme environments? Would this screening process work in our daily environments?
I think it is interesting that other strains have been isolated in other locations that have cold environments and it makes me wonder if there are strains that work similarly in hot to very hot environments.
How is Marine Oil Snow formed? Is it just an accumulation of oil particles that float back down from the surface of the spill? If so, is it possible to prevent this from forming by cleaning up the surface spill quicker?
Does the mineralization of hydrocarbon contaminants have any lasting effects on these environments? Is the product of this process not considered to be a contaminant or is it turned into a useful product for the environment?
I think it is referring to the controls that were killed with acid. But I do wonder why acid-killed controls were used rather than an E.coli strain as used in the last paper?
This was the comment I was looking for! The Meselson and Stahl experiments that we learned about in genetics were exactly what popped into my head when I read about heavy versus light DNA.
“Clearing”… is this similar to the Casein Hydrolysis test we did in Lab 5. Does this show the presence of caseinase exoenzyme production or something that has the ability to clear agar in the same way?
To add onto your question, I would also like to know if we have found this species or similar species in other ecosystems (like on land or in extreme conditions) and if they have the same roles in these areas or if maybe PAH degrading is something only the ocean-stricken species has adapted to?
How common is it for a microbial habitat to shift between an oxic and anoxic state in a 24 hour period? Is this similar to the day and night changes of plant photosynthesis and respiration?
I think it is interesting that this process has been found or studied in multiple very different sources: periphyton, which is located the vegetation found on the bottom of freshwater systems; the soil in Japan (assuming this is not from the sediment of a water system); and hot spring biofilms.
Would this anoxygenic photosynthetic As oxidation be common during algal blooms? Has this been studied?
I think this idea comes from haloarchaea being around for a long time, and the process of anoxygenic photosynthesis comes from early earth being anoxic.
I would be interested in further understanding the process of endpoint determination using liquid scintillation and why this was used to measure mineralization.
As with the last paper and although I do not fully understand these processes, I enjoy reading about real-life experiments that correlate with what we are doing in our own lab – I recognize those bacterial primers!
Does the color of these spring correlate to the abundance of each bacteria present?
What was the purpose of collecting samples months apart like this?
Yes, I think it may be dominated by Ectothiorhodospira because of the dominance of it’s ability to cycle arsenic, it may overpower any other organisms ability to cycle the arsenic.
Would the temperature range increases at night have anything to do with the respiration processes of other organisms in the environment?
I am just wondering why these various and specific locations for the soil, water, or sediment samples. I understand that the samples that were collected have various levels of salinity, because the bacteria needs to thrive in hypersaline environments as said in the introduction, but is there another property that the samples need to have in order to be used for their experiment?
The Lowry method sounded familiar, but I could not recall what exactly it was. So I googled it and found that the Lowry protein assay is a biochemical assay for determining the total level of protein in a solution. The total protein concentration is exhibited by a color change of the sample solution in proportion to protein concentration, which can then be measured using colorimetric techniques. This now makes sense as to why they used this method while using the UV spectrophotometer.
The fact that these archaea have been found to degrade both aliphatic and aromatic hydrocarbons in high-salinity environments leaves me to wonder if they can also degrade beneficial compounds to these type of environments? Or is it just exclusively degrading the hydrocarbons? What would be the cons to exposing these fragile environments to more bacteria and archaea?
I find the topic of bioremediation very interesting. The use of naturally occurring or introduced microorganisms to consume or break down environmental pollutants. This should be the first step when cleaning up pollutants instead of introducing more harmful chemicals in order to clean up other harmful hydrocarbons. This is especially important for the Gulf of Mexico. The Gulf has a relatively high number of oil rigs, while also being the home of many estuaries. This can be a dangerous combination.
[Approximately 40 to 60% of the added [14C]benzene was converted to 14CO2 in a period of 3 weeks (data not shown), further suggesting utilization of benzene as the carbon source.]
This is good news in the sense that over half of the added benzene was converted to CO2 in a period of three weeks. Which is relatively fast working in the world of science. I just need clarification on this part, as the strain Seminole degrades benzene, it is stated that benzene is converted into CO2. Does this mean as Seminole uptakes benzene, it releases CO2? Correct me if I am wrong, but could that just add more problems to these estuarine environments?
It is said that the pob and pca genes are found clustered together in a contiguous pattern or scattered over several portions of the genome. I wonder is it better for these genes to be clustered or scattered? Or does it make no difference to the expression of these genes? I happened to google it, but could not find any reliable information to go on
I am curious as to why the organism was not able to grow on catechol if it was predicted to be an intermediate in the benzene and toluene degradation pathway. I understand they didn’t find a conclusion for this, but is there a specific test to find out why? Or could it possibly be a mistake on their part when making predictions about the organism’s genome?
I would be interested in finding more out about the genes that they predicted would show during the degradation process, but didn’t. I am assuming once all the genes are well known, that is when we can start to build a plan towards degrading the compounds that are harmful to marine environments.
I am interested in learning more about fluorene degradation. Being in marine sciences, the main focus is usually carbon dioxide, methane, nitrogen, and other major greenhouse gases. People rarely think about other compounds that come into play as well. Then to learn about microbes that can degrade these toxic compounds and the science behind it is super interesting,
I looked up the bacterial species as well and found that there are 8 halophilic and 10 halotolerant different strains for this particular bacteria. Im curious to see any similarities with the last degrading halotolerant bacteria we discussed for paper 1
I was wondering why they mentioned that they used phosphate buffer instead of Tris buffer. So I looked up the difference of the two that might lead as to why they choose one over another. Tris buffer is used as a running buffer in agarose gel electrophoresis to identify a species by looking at the base pairs. On the other hand, phosphate buffer is used in isolating cells from tissue to maintain pH and keep cells alive. I also saw where phosphate is more expensive, but I see why it was needed instead of the Tris buffer for this experiment.
I like how detailed these authors are with their experiment. It makes it easier to read and understand when they explain each step and what the product resulted after doing each step and not just assuming the reader knows how they got to a specific result.
Coming from a now second time reader of scientific papers.
Looking at figure 2, does this mean that the Terrabacter sp. DBF63 is the species found to degrade fluorene more successfully compared to the others?
I too would like more explanation on this. I am also confused on the importance of the functional expression of FlnA1 and FlnA2
I think it is interesting how we are now on our third paper about degradation of contaminants in the marine environment by naturally occurring or introduced organisms, and I have never heard anything about it beforehand. Introducing these organisms to polluted areas seems like such a good idea. I wonder if this is actually taking affect anywhere around the world
Since fluorene had three different degradation pathways, I wonder if fluoranthene will have similar pathways
I didn’t know about the cre-lox recombination method so I did a little research. Cre-lox recombination involves the targeting of a specific sequence of DNA and splicing it with the help of an enzyme called cre recombinase. Cre-lox recombination is commonly used t circumvent embryonic lethality caused by systematic inactivation of many genes.
I think it is interesting that they took samples from the Indian Ocean sediment, but then grew the strain on artificial sea water. If this is to work in the marine environment, wouldn’t it be more beneficial to take ocean water samples as well instead of something man made?
Could you further explain this figure? There is so much small information, I am not sure what to take out of the circular map. I am also confused to what the inner circle is showing
This is the most genes involved in the degradation of PAHs in a single strain that we have read about. Does this mean that the P73 is more capable of degradation of hydrocarbons than any of the other strains that we have learned about in the previous papers?
Why don’t we know about the degradation of PAHs with higher molecular weights? Is it due to the large size of those PAHs?
How does naphthalene hinder mitochondrial resiration?
The variety and number of tests performed should show relationships among naphthalene degrading bacteria. Are there any other tests that would help with the study? Is there any particular test that is more beneficial or conclusive in determining naphthalene degrading bacteria?
ONR7a medium was used since it contains naphthalene as the only carbon source. Are there other media that have similar characteristics?
Since strains N2, N7, N9, and N10 are all phylogenetically related (from figure 2), one would assume their growth rates would be nearly the same; however, the growth rates range from 0.345 to 0.928 (from table 1). Why is their so much variation?
Why is it that 400 ppm was the optimum concentration of naphthalene? Is it more important simply to understand that 400 ppm is the optimum concentration of naphthalene?
I find it interesting that in paragraph 4 gram positive bacteria were isolated and found to play a role in naphthalene degradation, but in this paragraph, gram negative bacteria were favored for naphthalene degradation. Does the difference have to do with the lipopolysaccharides in the gram negative bacteria’s outer membrane?
Since bacteria were isolated from an oil polluted area in the Persian Gulf, could bacteria be isolated from the Gulf of Mexico which was polluted during the 2010 BP oil spill? I think it would be interesting to isolate bacteria from a waterway that most people in this region have encountered.
What is the largest aromatic compound ring number that bacteria are able to degrade? Is this dependent on the type/complexity of the bacteria?
I know some of the main points of the study were to isolate and classify naphthalene degrading bacteria, but unless I missed it, the paper didn’t discuss how to insert these degrading bacteria into oil polluted environments for controlled bioremediation processes. Is this being tested?
Are there any preventative measures to see if the seafood we are consuming has toxic PAHs in it?
Where are these Cycloclasticus bacteria located? Are they in any marine environment polluted by petroleum?
Various chromatography techniques were performed in the method of this lab. Chromatography is useful in separation by size, charge, and affinity each of which is useful in identifying a pure isolate.
Various chromatography techniques were performed in the method of this lab. Chromatography is useful in separation by size, charge, and affinity each of which is useful in characterizing a pure isolate.
Since naphthalene is a two ringed structure and phenanthrene is a three ringed structure, is it important to know which bacteria can use which chemical? Or is the fact that they have the ability to degrade either and/or both of the compounds the topic of interest?
Why are E. coli containing pPhnA able to transform certain molecules but not others?
All of the aromatic substrates were hydroxylated in the figure. Is this useful for them to be degraded?
The Sau3AI fragment of DNA is the gene which allows pH1a and pH1b to oxidize certain aromatic compounds. Is this inerpretation of the paragraph correct?
What is the significance of not finding the gene clusters for PAH degradation on the plasmid, and why doesn’t the strain have a plasmid at all? This is a unique characteristic as plasmids often contain genes for antibiotic resistance, degradation properties, etc.
Monooxygenases are enzymes that transport one oxygen atom from the air to the substrate, and dioxygenases are enzymes that transport both oxygen atoms from the air to the substrate. I think this comparison is useful if, like me, you have trouble keeping these definitions straight.
If other dioxygenase genes in Cycloclasticus were examined, what would those results conclude? Since this paper is several years old, I assume additional research has been done.
Is there another method for obtaining the aresenite oxidase gene since the PCR for the genes was unable to be obtained?
How are some organisms able to utilize arsenic, but for others arsenic is toxic? I understand that if an organism can utilize arsenic it does so by using it as an electron donor or acceptor, but what determines whether or not an organism can use arsenic?
Alkalithermophiles are extremophiles that are grow optimally at high pH (around pH of 10). The rod-shaped bacteria, Anaerobranca californiensis, is an example of an alkalithermophile. From further research, I found that this bacteria grows best in the presence of elemental sulfur, polysulfide, or thiosulfate which are found in the spring waters.
A radioassay tests a radioactive sample to determine the intensity of its radiation. Incubation is a process that maintains a favorable temperature and other conditions promoting development. In this experiment, is it correct to deduce that radioassay incubation was used to “grow” the bacteria?
Assimilative reduction is where an element is consumed and incorporated into new cell material. In light-incubated slurries, assimilation was not stimulated by the presence of arsenic oxyanions. Why is that?
Are there further tests that could be performed to prove that the clones were of the genus Ectothiorhodospira, or are the results obtained sufficient?
Senescence or biological aging is the gradual deterioration of function characteristic of most complex lifeforms. Essentially the third sentence is saying growth rates decrease because the microbes are aging and toxins are accumulating.
Is there any way to make a guess on which microbes reduced arsenate?
Sulfide and hydrogen increase the rates of arsenate reduction because they are at the top of the standard reduction potential chart. This means they give off a lot of energy.
Where do the microbes get the bicarbonate?
Fluoranthene is a fusion of naphthalene and benzene by a 5 member ring. An alternate PAH is one where the pi centers are not adjacent to another pi center. In a non-alternate PAH, somewhere on the ring there are pi centers that are adjacent to one another.
From further reading of the paper we will find the answer to the following question, but for now, could it be guessed that the fluoranthene degradation pathway of Celeribacter indicus is initiated by dioxygenation as said in paragraph 2?
One of the goals of this experiment was to isolate the genes needed for PAH degradation. Should genome sequencing help to determine the fluoranthene degradation pathway?
I had never heard of the term silylation, so after a quick google search, I found that this is replacing a proton with a trialkylsilyl group. The purpose of this is for analysis using GC and mass spec. How is this helpful in our understanding of the fluoranthene degradation pathway?
If glucose-6-phosphatase is missing, how does gluconeogenesis proceed? Were these genes not transferred during horizontal gene transfer?
I’m having a difficult time understanding why strain B30 and strain P73T were compared. Are the two strains more or less the same other than B30 lacks most of the RHD genes found in P73T?
Are there other genes throughout the genome that could also be responsible for the integration of foreign sequences needed for HGT? In this paragraph it talks about region B, but what about regions A, C, and D?
Knocking out a gene is a good way to determine if a gene has a suspected function. In this approach, gene function is studied by examining gene loss. What other methods can one use to determine a gene’s function? Genomic DNA analysis?
Why is aromatic ring hydroxylation the most difficult step in fluoranthene degradation? Is it a difficult reaction to get started, does it make unnecessary side products/reactions, or is it something completely different?
Horizontal gene transfer is a good way for new genes with new functions to be inserted into a genome, but can HGT be harmful to an organism? Can it insert a gene that inhibits a necessary gene? I assume such organisms wouldn’t survive in nature since this would decrease the organisms’ fitness.
Sphingomonas sp. strain LB126 is unique from other microbes we’ve looked at since it was isolated from PAH-contaminated soil. We usually look at microbes from PAH-contaminated marine environments. Is there any difference among microbes found in soil vs in a marine environment?
Why is there more research on fluorene degradation of gram-positive bacteria than gram-negative bacteria?
I did some googling on degenerate primers. I found that they’re useful when isolating the same gene from several organisms, as the genes are most likely similar but not identical to one another. Degenerate primers are also used when primer design is based on protein sequence.
From my biochemistry course last semester, I learned that SDS-PAGE is used when separating proteins by electrophoresis.
From what I found, sonication involves applying sound energy to agitate particles in a sample. Some of the purposes include: to break intermolecular interactions, for the production of nanoparticles, to disrupt or deactivate a biological material, to evenly disperse nanoparticles in liquids, initiate crystillization, loosening particles adhering to surfaces, etc.
For this experiment, is sonication used to break up the pellet into the liquid?
Is this because they wanted to obtain a clone with a site for one enzyme on one end and a site for another enzyme on the other end?
I’m trying to make sense of the information found in paragraphs 6-8 and in Table 2. From what I understand, this is to compare what substrates FlnA1-FlnA2 can use to which substrates two common dioxygenases, DFDO and CARDO can use. Are FlnA1-FlnA2 genes that were found in Sphingomonas’ genome that function in fluorene degradation? From these 3 paragraphs and table 2, what exactly should we find to be noteworthy?
I think it is very interesting that a gene usually only found in gram positive bacteria is found in Sphingomonas, a gram negative bacteria.
Additionally, I think this paragraph helps to clear up some of the questions I had about FlnA1-FlnA2.
Cells use glucose in glycolysis to produce pyruvate. Pyruvate can then be broken down to lactate in lactic acid fermentation, ethanol in ethanol fermentation, or acetyl CoA in the citric acid cycle.
Shouldn’t we expect that fluorene is not the best carbon source since smaller PAHs such as naphthalene are easier to degrade? Or am I confusing this with the fact that smaller PAHs are more widely studied as they’re model PAHs?
Is it unique that Sphingomonas sp. strain A4 is only able to grow on acenaphthene & acenaphthylene and not lower molecular weight PAHs?
I looked up the 16 priority PAHs. From what I found, these PAHs are of high priority because of their potential toxicity in humans & other organisms and their prevalence & persistence in the environment.
Is this describing targeted mutagenesis like we went over in class today?
From what I found on google glass power is used for purifying fragments from agarose gel electrophoresis, is that correct? What is the purpose of using glass powder?
Blunting of sticky ends is done to allow non-compatible ends to be joined.
Shotgun cloning is different than shotgun sequencing. Shotgun cloning is used to duplicate gDNA by fragmenting DNA with a restriction enzyme. These fragments are then cloned into a vector. In shotgun sequencing, DNA is randomly fragmented with a restriction enzyme, but then, the fragments are sequenced. These sequences are reassembled on the basis of their overlapping regions.
In this paragraph, they used targeted mutagenesis which relies on homologous recombination. I’m going to attempt to explain it, so please let me know where I explain it incorrectly. In this type of mutagenesis, the arhA1 gene was cloned into a suicide vector. The antibiotic resistance gene (Gm) is inserted into the arhA1 gene (in the vector) which disrupts its function of PAH degradation. Homologous recombination occurs, so now the mutated arhA1 gene is inserted. This allows the gene to show antibiotic resistance, but not PAH degradation. You know you’ve successfully mutated the gene when the original function is inhibited.
Why was the phylogenetic tree constructed like this? Is this advantageous to the trees we’ve made?
Have the additional studies mentioned been done to show other genes needed in the degradation of acenaphthene and acenapthylene?
So in this experiment, genes encoding for ferredoxin & ferredoxin reductase were on one of the plasmids that was transformed in E. coli. I know ferredoxin works in electron transfer, but what exactly is happening with it in this experiment? What is the purpose of having a plasmid with genes encoding for ferredoxin and ferredoxin reductase?
Transposon mutagenesis allows genes to be moved to the chromosome of your organism of interest. This causes mutation because the gene is inserted in the middle of a functioning gene. In this experiment, they’ll mutate the genes involved with acenaphthene degradation.
In paper 3, the authors found the genes involved in the initial steps of acenaphthene degradation, oxygenation. In this paper, the authors hope to find genes involved in the next steps of acenaphthene degradation and the genes involved with acenaphthene degradation regulation. Would sequencing the organism’s genome give you this information?
I tried to do some googling on what a xenobiotic is. From what I found, they’re substances which are foreign to the organism. Xenobiotics include drugs, industrial chemicals, naturally occuring poison, and environmental pollutants. This website might help to answer some of your questions.
http://www.ilocis.org/documents/chpt33e.htm
I know that sometimes the order of events in the papers is not always written in the way they were carried out. Is that the case here after the DNA manipulations paragraph?
I’m assuming arhR is a gene involved in acenaphthene degradation. Disrupting the gene will give us insight into the function of the arhR gene. From this paragraph, are we able to determine what disrupting the gene caused to happen, or would this information be found in the results section?
Does the location of the insert tell us that acenaphthene degradation is dependent on these genes?
I was unfamiliar with what a primer extension analysis was. From my google search I found the following:
Primer extension allows the 5′ ends of RNA to be mapped. It can be used to determine the start site of transcription by using a primer which is radiolabeled.
A transposase gene is often a sign that horizontal gene transfer occurred, right? Is that the case here?
I’m not sure how old this paper is, but has further investigation of the degradative genes and regulatory mechanisms been done to help clarify the acenaphthene degradation pathway?
Is there a way to place an insert into a particular spot in an organism’s genome? If not, is that why there were 10,000 strains with inserts?
After the process of bioremediation, what type of effect does the end product of microbial biomass have on the environment? Does it show an effect at all?
What are the costs and effects of using different strains of bacteria for biodegradation? What difference would you expect to see when deciding between bacteria based on their biodegradation ability?
What is the purpose of transporting the samples on ice? Is there a certain temperature that is required to maintain?
What is the CTAB method and how does it work?
What is the GC-FID method and how does it differ from spectrometry?
What factor would determine the amount of growth of strains after 400 ppm concentration?
Would Gram-negative naphthalene bacteria be able to play as great a role as Gram-positive in an oil-contaminated area? Or is that possible?
Is it possible for Gram positive bacteria to be found in an area of high pollution of heavy crude oil with Gram negative bacteria?
What other gene sequences besides C23O are most useful in identifying phylogeny of a strain?
By what measurement determines which bacterial strain is best suited for metabolizing PAHs?
I read that sonication can cause cell lysis. But in this case, is it used to just agitate the cells?
I looked up SalI, but I am still unsure of exactly what it is and what it does?
Looking at the data table, I am unsure of exactly what the numbers corresponding to each carbon substrate represent? I understand the table shows the utilization of each carbon substrate but I am unsure of how the numbers are calculated.
Is the basic 7.0 pH level usually the ideal when it comes to the strain degrading PAHs?
How would evolutionary pressures cause species of Pseudomonas putida to be scattered to different clusters and those from Sphingomonas to be in the same group?
How does the structure of the meta-pathway operon change the way S. paucimobilis degrades PAHs?
What would cause an inability to obtain PCR products for axoB genes?
Would bacteria using arsenate as a respiratory electron acceptor benefit more than bacteria using arsenite as an electron donor?
What is the significance in the colorization of the different bacteria? Is green always associated with cyanobacteria?
Is there a particular reason all aoxB clones failed to show any similarity to the database?
Why would the re-introduction of light cause a diminished rate of As (III) oxidation?
Since the promotion of O2 exchange didn’t affect the rates, what can be concluded?
Would there be a method to more directly determine which biofilm microbes were associated with these arrA amplicons involved with As(V) reduction in dark incubations?
My guess to your question based off the section is that V. fischeri ES114 contained something that the others did not which inhibited the growth specifically to that culture, because it was stated that the other culture fluids from the other tested strains did not inhibit V. harveyi growth.
I also noticed how well of a specimen vibrio seemed for the experiment, knowing this information makes me think the results of this study will be great in terms how much data they will be able to collect.
Reading the paragraph right above this one, siderophores have a high affinity for Ferric or Fe^3+ iron which is the abundant form of iron in at neural pH in the presence of oxygen, so I came to the conclusion that if it is not that specific affinity for iron then it would not actually bind. In conclusion not all siderophore producing microbes would not have the same affinity to iron.
Oops! This comment was supposed to be in response to Ryan.
I actually knew that nitrification played a role in marine life, but I was not aware of the significant role that it played until I just read the data. So, I also agree that this was a pretty interesting finding.
After reading your comment, I was curious and decided to research your question. From the looks of it, as if right now, they are not able to give too much more information other than that diversity is negatively impacted. So, I am guessing some life exists but not a lot.
Based on your definition and the paper’s definition of what a steady flux is, it seems to a constant considering it is distinguished by the amount of atoms crossing.
I think they were determined or at least thought to have made an appearance in the step above where it says “Anammox rates were measured by the means of isotope-pairing” and were then verified using the CARD-FISH.
“Hydrocarbon contamination in Antarctica has profound effects that have been shown to reshape the structure of microbial communities as well as affecting the abundance of small invertebrate organisms”. How exactly are the microbial communities reshaped? What is to be expected of the communities when they are encountered? Also, does that mean there is a reduced amount of small invertebrate organisms or more of a diversity when taking into account their amount?
In response to your question regarding limiting the amount of contamination, we would have to figure out what might be one of the biggest contributions in causing the contamination in order to reduce it. Because the causes of oil contamination could easily very. If that was done, then I feel confident that we could play a role in the reduction of contamination.
Finding your question interesting as to why biofilm may not occur in this experiment, I researched it. Based on my research, the formation of biofilm has to do with antibiotics, biocides, and ion coating. Those can interfere with the attachment stage in biofilm which will cause the other stages not to necessarily be applied.
I also wondered what the change in temperature would do to the buffer. Personally, I think it would make the 90 minute incubation drop, but what will the changes in temperature do to the results of the experiment? what direction will it go?
[ To this end, we assessed the ability of culture supernatants to emulsify diesel (E24 index) as well as the ability of supernatants to generate water droplet collapse by altering surface tension (Figure 3G). In both tests, D43FB supernatants lacked emulsifying abilities, and exhibited similar behaviors to negative controls, suggesting D43FB does not secrete biosurfactants to aid in its degradation of phenanthrene.]
While reading, I was not necessarily sure what “emulsify” or “emulsifying” meant. I researched the terms and they basically mean to have two liquids in a colloid and not completely mixed together. So, in other words, it now makes sense as to why emulsification had nothing to do with the degradation of phenanthrene due to the lack of emulsifying abilities.
I agree with you as far as not truly believing the D43FB will thrive when put into the conditions of Antartica. It seems as if they are more so of hoping for the results will turn out good solely because it had the best results when it came to the others, but the analyzation doesn’t seem too solid.
In addition to what you’re saying Ryan, I would also love to compare how S. xenophagum D43FB would respond in the situation in which the temperatures change drastically either much colder or warmer. Does the weather play a major role in how the isolate degrades and replicate?
I was not aware until reading the article that it was forbidden to bring foreign organisms into Antartica, so I was caught by surprise as well. Also, I feel like this should be enforced in a lot more other places as well because according to the data, strict laws such as this are what is making this environment such a great place. There are so many places that could benefit from the narrowing down of decontamination techniques/strategies.
My initial thoughts after reading was what necessarily makes iron a great source of competition as opposed to any other source and what lead them into making the decision as to why they should use it?
I would assume that there would be a larger possibility to make a mistake considering how broad the approach is rather than specific.
In response to your question, I do not necessarily think it shows evidence of that because of it stating the V. fischeri ES114 does not produce aerobactin and/or sufficient iron. In other words, it doesn’t clearly state if low iron is the sole reason.
I think it’s very interesting to see the drastic change when the oxygen has nicely depicted lines decreasing into the suboxic area then travels backwards increasing back into the lower oxic area. Then the H2S starts off at a plateu or a completely flat rate then gradually increases as O2 completely runs out.
I am a little confused in Figure 2D because what I am interpreting based on data is that the NOx- decreases in some areas of the lower oxic area while it slightly increases then reaches a rate where it plateaus and does nothing at all like it was at the beginning where there was no oxygen? So, is this saying that the oxygen ran out?
I definitely agree with your comment, without the genes being transferred in that manner, the experiment would not be as effective. I wasn’t aware of the horizontal transfer being across multiple lineages either until reading this section of the article.
Yes, this last sentence gave a great/more clear summary of what was actually concluded at the end of the experiment. Before reading it, I did have some confusion of what they were suggesting. I found this experiment interesting because I have never seen this type of interaction “occur in nature”.
I honestly don’t think it would be considering the genes were rarely seen to begin with, so I think it just happened to be abundant in that particular location based on the conditions. The only way I feel like would be observed is if they had similar conditions.
I would actually like to know the answer to that question as well, because I thought they initially chose the Black Sea due to its great source of microaerobic nitrification. If I had to guess though I would say that it would not be as directly coupled which it what would make the Black Sea so unique to begin with.
I found bioaugmentation very interesting. I found a study where they actually used bioaugmentation and biostimulation hand in hand and discovered that together these two processes were optimal for oil degrading. Interestingly, there are Antarctic bacteria that are capable of hydrocarbon degradation at extremely low temperatures and they concluded that the use of bioaugmentation in those situations could enhance the rate of bioremediation.
I found myself pondering similar things during this reading. My initial thought was that it would be doing more harm than good as far as bringing those pollutants over as well as any non-native species of microbes and things that are on or in the icebergs. I typically tend to believe that disrupting the natural order of things means it shouldn’t happen. However, with what I’ve learned about serious drought issues that people are facing I feel this is a fairly logical route to supply the people. The captain who conceived the plan seems to have an ecological, well- thought out plan to make it work. I think the pollutants would be in such minimal amounts that purifying that would become the easiest part of the process.
I don’t necessarily believe the experiment would have been hindered if another media was chosen that suppressed certain bacterial growth as long as there were runs made with a control media. However, this is obviously very dependent on which media is used.
When pondering which media (if any) I though would have been more successful, I found myself thinking that R2A is typically used with slower growing bacteria and can be often suppressed by faster growing colonies. This made me wonder if they came across this issue in the field. Unfortunately, I never really found the answer for which agar would be better than R2A. I would lve to hear others thoughts.
After reading about biofilm production as well as this paragraph, I was curious as to which of the 350 colonies produced were the ones producing the biofilm and if there is a method to differentiate between those working in biofilm production and the colonies who didn’t. If it wasn’t all of them.
I believe the answer is yes! When undergoing transformation, the cell lyses and releases those intracellular components into the environment. That includes DNA fragments which a bacterium could take up to gain resistance.
I was also confused. From what I understood when looking it up, I found that while most siderophores are strong enough to pull iron from host-binding proteins so maybe since the microorganisms producing them are in iron deficient areas its just another case of survival of the fittest we observe in countless environments. There are siderophores who outcompete others based on better genetics.
From what I looked into I understand it to be a more specific type of PCR (DOP-PCR). It allows the employment of olignucleotides of partially degenerate sequences specific for genome mapping. Olignucleotides primary use seem to be primers for PCR so your assumption is correct. As for specific reasoning, I found they have the ability to amplify a small amount of DNA as well as being designed for length and G/C content. They flank the target reason so it seems just to be a wonderful primer for this use.
I read that this type of assay seems to be the most accurate as compared to counting-based methods for plotting growth curves. Is this because the fluorescence- based method isn’t dependent on determination of total cell numbers, but more so assessing fluorescence of the given sample of single cell from the population at a given time? I feel that contributes to lower variation rates among the plated samples which in turn only increases reliability especially when assessing the lag, log, and stationary phases.
I understood it to be both. They kind of go hand in hand as you can predict antibiotics resistance based off of mutations. Mutant construction of essential genes aid in resistance. Or am I missing some context?
Specifically CAS because of its ability to detect various types of siderophores. I found a paper that was saying in comparison of solid versus liquid that the liquid reaction-rate was an positive over the solid in all species tested. So maybe it was easier to quantify this way?
[The inability to grow was not specific to V. harveyi: V. fischeri ES114 culture fluids also prevented growth of Photobacterium angustum S14 and Vibrio cholerae C6706, while no diminishment of growth yield of Vibrio parahaemolyticus BB22OP and Vibrio vulnificus ATCC 29306 occurred (Fig. 1B). ]
comparing this to figure 1B made for much easier understanding. However, I found it a bit confusing that the growth inhibition was specific to V fischeri ES114 while others still had the inability to grow. If someone could offer some clarification.
[In communities, public goods can be exploited by cheaters who acquire advantages through use of the good but who do not pay the energetic cost of goods production. In the context of siderophores, a cheater need only possess genes required for recognition and uptake of the siderophore-iron (Fe3+) complex. ]
This comparison helped my understanding greatly. I found it very interesting that the cluster of siderophore biosynthetic genes encode for the ancillary functions of reception and transport for the V. fischeri.
I know that this just saying that V. fischeri ES114 outcompetes the others by uptaking all the iron which in turn also deprives the other of iron. Would the cheaters not steal the siderophores to outcompete V.fisceri ES114 since it wouldn’t be paying the metabolic cost to produce/secrete?
[ This co-culture system, in which either or both species can be genetically manipulated, provides a route to the quantitative investigation of both competitive and cooperative interspecies interactions that occur in nature.]
The identification of aerobactin acts as an inhibitor or the V. harveyi while working towards eliminating production in the V. fischeri ES114 thus creating this co-culture. I found this so interesting and somehow it helped the whole discussion click easier for me. It’s cool that this experiment led to quantitative data for the competitive and cooperative interspecies interactions.
[ nonthermophilic MGI Crenarchaeota constitute a significant portion of oceanic picoplankton (up to 30%) (21, 22) and a considerable fraction are likely autotrophic (23, 24), it is speculated that these MGI Crenarchaeota could be more important nitrifiers in the oceans than the usually less abundant AOB (18, 19). ]
As a student who has been studying marine science, I found this particularly interesting specifically for its higher abundance. Nitrification in oceans is important to the nitrogen cycle. For several reasons, including the microorganisms that use it for nutrients. I am eager to read where this study went and infer the impact it will have on things I’ve learned this far.
I think nitrate levels increase with depth because there’s no producers consuming it as well as regeneration due to decomposition of organics and the hydrothermal vent systems.
I looked up what a steady state flux is and from what I read its a diffusion process distinguished by amount of atoms crossing a unit of area perpendicular to a given direction per unit of time. So basically it is a constant concentration gradient?
I noticed they got their water samples and examined physical properties. Chemical analyses, dark carbon fixation, and Anammox rates were measured. Presumably, this data could be used to provide evidence of the amoA activities in the water column from which the samples were taken. This also provides comparative data for the coupling between the nitrification process and anammox along side the chemical profiling.
In Fig. 1, I would say these data points would be expected. In part a it shows nitrogen levels tend to be higher/largely distributed in greater depths due to lack of light and producers as well as regeneration through decomposition of organics by bacterial colonies.
Part b: Oxygen is most abundant in surface waters and decreases with depth. Sulfide would increase with depth as we get deposits of decomposing matter settling down.
Part c: light transmission is obviously one that is higher in surface waters and dissipates with depth as the light is scattered or absorbed by particles floating in the water. The particles increase with depth and in return decrease light penetration.
Part d: Anammox bacterial cells are more abundant with depth as they love abundant ammonium and nitrite as well as dark, anaerobic conditions (which matches the graphs before). The NH4 spikes where there arent more dense colonies and decreases when bacterial densities increase.
Fig. 4 shows that the most efficient production of 14N15N and 15N15N are converted from 15NH4 (a) where both numbers steadily increase.
(b) From 15NO2, we get equal 14N15N, however, 15N15N was not produced.
(c)Finally, with 15NH4 + 14NO2, we see 15NO2- produced to equal amounts, but drop off. And 14N15N had steady production, but the numbers weren’t quite there. Also showing no production of 15N15N.
[In accord with previous findings, dissolved oxygen in the central Black Sea (43°14.9′N, 34°00.0′E) [supporting information (SI) Fig. 5] decreased from fully oxic to <5 μM at 85 m (σt = 15.83, for comparison with studies in other parts of the basin) (Fig. 1). The suboxic zone extended from this depth to 112 m (σt = 16.15) below which sulfide started to accumulate. ]
I was expecting a similar discussion to this based off of just the graphs, however, I was not as well spoken. Although I falsely assumed the graph down to an anoxic level instead of just a suboxic zone. The sulfide was predicted to increase in the suboxic zone because of the sulfur-reducing bacteria and their tendency to thrive in a lower oxygen environment.
[ The total NOx − production in the oxic zone (55.5 μmol of N m−2 day−1) also could not match this NH4 + loss because of anammox, which consumes 1 mol of NO2 − per mol of NH4 + oxidized. Therefore, an additional local source of NO2 − and/or an additional loss of NH4 + must be present to reconcile the difference. ¶ 16 Leave a comment on paragraph 16 0 The best candidate to explain this phenomenon is microaerobic or anaerobic nitrification, whose direct coupling with anammox, the so-called completely autotrophic nitrogen removal over nitrite (CANON), has been demonstrated in bioreactors (35, 36). At 100 m, nitrification was evidenced by the production of 15NO2 − (12.9 nM day−1) in incubations with 15NH4 + + 14NO2 − and no measurable oxygen (Fig. 4).]
The coupling of anammox and microaerobic or anaerobic nitrification was apparent when analyzing anammox rate measurements and the isotope pairings in Fig. 4. I just found this part of the experiment particularly interesting because it gave me more insight into whether NO2- in marine water columns comes from the nitrification process, nitrate reduction, or both. Directly ties into my studies in physical oceanography.
Good question Justin, I was thinking the same thing. I think that it does generally occur when smoke enters the air, rather it be burning firewood or smoking a cigarette. PAHs are found in a various amount of things that we tend to burn, therefore I feel a common way they get exposed to our environment is through smoke in the air.
I agree with you Dawson! I find it intriguing myself how much science and new technology have advanced allowing us to isolate and identify one bacterium from another. I find it interesting that we can isolate these enzymes and further manipulate them to see what they can do.
Why is it that more information is available on bacterial biodegradation of lower molecular weigh than of high molecular weight? What factors play a role in performing microbial degradation? Are these processes also harmful to the environment?
Is the Persian Gulf still suffering from being polluted in 1990? How does the PAH bioremediation process converge the pollutant material into a useable substance that is not harmful to the environment? Can the marine environment actually benefit from these strategies?
Why did the samples have to be incubated for a seven day period? How could they actually determine the amount of growth if the samples weren’t being directly measured?
What does the action ” agitating in the vortex” mean? How does this process of using gas chromatography work? How long does it take? Is this the most accurate way to calculate the culture medium?
What caused the strains show a dramatic decrease in growth after 400 ppm concentration? Why did the Salegentibacter sp. strain N7 have the maximum growth.
What is the process on how the activity and adhesion were examined? What was different about strain N1 that allowed it to have the highest values of emulsification activity and values of cell surface hydrophobicity?
What is a catechol? How would they be able catalyze? Why is catechol 2,3- dioxygenase the most important member of the meta-cleavage pathways?
What does the 16S rDNA sequence control? Why was it necessary for them to clone the catechol meta-pathway operon genes? What characterizations did the strain contain that was necessary for the conclusion that it is a good model organism?
What is the difference between the BHY plate from the plates we use in lab such as MSM and R2A? What type of plate is the BHY plate?
Why was it necessary for them to disrupt the cells? What would happen if the cells were not disrupted? How does the centrifuge remove the cell debris?
What other organisms belong to the genus Sphingomonas? Did all of the results show that the strain should be apart of this genus or were there specific traits that made them think this?
Why did the strain ZX4 oxidize only 55 out of the 95 carbon sources? What does the paucimobilis describe?
Is there any relationship with the distance of spacing between encoding genes? When it says the arrangements are similar to others, does it mean they function the same?
What is the ideal or preferred structure of the meta-pathway operon? How do the differences in structure alternate the understanding of metabolic capacities?
Are there specific advantages in being oxic, anoic, or shifting between the two?
Why weren’t they able to obtain authentic PCR products for the arsenite oxidase genes? Was there an error? Would they normally have been able to obtain this information?
Why did they take more samples and focus most of their attention on the samples that were taken from the red springs instead of the green colored ones?
Why did they use a toothbrush to remove the biofilm first? Then used a spatula to remove the biofilm for the DNA analysis?
When the light was reintroduced, why was the rate of oxidation lower than when it was introduced the first time.
Did they make intentionally want the temperatures to range, or was that one of their findings?
Are they saying that since the biofilms have already been exposed to variations in the environment and have already been introduced to sulfide and hydrogen, that they did not respond drastically to the addition of sulfide and hydrogen that they added?
Was there a specific reason that they decided to use sulfide and hydrogen in their study? Why didn’t they use any of the other elements?
What is the function of a benzene ring? So the more rings it has the harder it is to break them down?
Why does the Persian Gulf have such a high annual oil contamination? Has this increased or decreased recently? Are there any other places that have higher numbers of oil contamination?
Since the bacteria have different decomposing enzymes, is it possible to combine multiple bioremediation methods to degrade PAHs with higher molecular weights? If these methods were combined, is it possible that they could have an inverse effect & end up doing more harm than good?
Since PAH bioremediation is effective and environmentally benign, are they being done on large scales? Are the residents of the area accepting of the bioremediation, or are they doubtful and afraid of the science behind the treatment?
Is there a reason why the culture was incubated just for 7 days?
Were the phenotypically different colonies placed on the fresh ONR7a agar together or were they separated according to different size requirements?
Is there a reason why the strains were incubated just for 15 days? Could they have endured a longer incubation period?
These numbers have a lot of variation. Could it be due to some of the strains dying off and some of them regenerating?
Were the seawater samples taken in the same area? Could differences of salinity of the seawater samples effect the number of naphthalene-degrading bacteria strains collected?
Although this form of bioremediation is helpful for marine ecosystems in the Persian Gulf, can it be helpful in more rural marine areas as well? Could the rural environment alter the type and number of naphthalene-degrading bacteria?
Why is the 16S rDNA sequence being used instead of other DNA markers?
Are there trade-offs for being able to degrade PAHs rapidly?
Bushnell-Haas minimal salt medium is ideal for studying the microbial degradation of hydrocarbons and examining microbial contamination for fuels.
Luria-Bertani medium is ideal for bacterial growth because it’s rich in nutrition.
Both of these mediums seem like good choices to produce phenanthrene cultures.
I found that Glutathione S-tranferase are detoxifying enzymes, which may help with the biodegradation of phenanthrene.
Since the non-inoculated media are used as references, are they also considered the negative control?
I found that trichloromethane can be used for oil extractions and the purification of antibiotics.
Although, Sphingomonas is a naturally metabolic bacteria that is very versatile and works well in bioremediation, some of its species can be dangerous for some organisms. It seems that its versatility could be a trade-off for safety.
Although 16S rDNA sequences have wide ranges of information about bacteria, could other genetic markers be used as well?
Although Sphingomonos is versatile and biodegrades PAHs well, it is corrosive to copper pipes. This could undo the progress of the bioremediation and possibly effect neighboring water supplies.
GSTs can be used to detoxify xenobiotic cells, which is good in some organisms, but they have the chance of causing resistance. They can resistance in chemotherapy. Is it possible that they can form resistance in the biodegradation of phenanthrene and other PAHs as well?
Periphyton contains a mixture of autotrophic and heterotrophic aquatic microbes, including cyanobacteria.
Halobacteriacaea are able to survive extreme environments such as the hot islands of Paoha Island.
Ectothiorhodospira are purple sulfur bacteria that can be found in anaerobic, marine ecosystems.
HPLC works fast, yet it is able to separate small particles efficiently.
If there were environmental variables, such as air pollution, would that need to be accounted for in the artificial medium as well?
Photosynthesis occurs during the light-incubation, using arsenite as the electron donor and arsenite as the electron acceptor.
As(III) and As (V) have inverse relationships due to being incubated in the light or in darkness. Therefore, as one increases, the other decreases.
Photosynthesis drives the light reaction, which decreases As(III). The chemolithotrophic process drives the dark reaction, which increases As(V).
Since sulfide was the more readily available electron donor than H2 in the oxidizing step, the H2 levels dropped.
While As(V) reduced to As(III), H2 donated as many electrons as it could and sulfide rates increases.
Would a weaker terminal substrate work better than acetate?
I found it interesting that both the Ectothiorhodospira and the uncultured both had the highest identity percentages, although the Archean seems to be unidentified.
I can understand how PAH exposure through respiratory uptake can cause harmful effects. I’m curious whether or not the dermal uptake is present in the same situations where respiratory uptake is occurring. I would guess higher concentrations of PAH would be required for dermal uptake.
Are inorganic nutrients able to provide energy in order to power the metabolic pathways used to degrade PAH? This could be be accomplished by harvesting energy from redox reactions or possibly promoting an excess of reactants forcing the reaction to move forward.
It would be interesting to see if the bacteria of interest in this study could be supplemented by certain nutrients to target Phe instead of Fla. After reading the entire study, it seems like this could be a possibility.
I wonder if pH 7 is the optimal growing condition for most of the cultures used throughout the experiment or if it was selected for simplicity purposes. Keeping it consistent throughout the experiment is a good standard but I wonder how it affects the growth of various bacterias.
Extractions were commonly done throughout organic lab. It’s interesting to learn about an example in which this lab technique is relevant to microbiology!
This experiment seemed to be a little bit of a side track. Was the purpose of this experiment just to compare how this bacteria degrades ILCO as opposed to other strains that had already been studied.
It’s very interesting how the rate of degradation is not linear. I would have expected a linear line or even a line demonstrating exponential decay. There’s also a fairly large standard deviation on the R. erythropolis in MSM for a few values that may distort the line.
It’s very cool in this experiment that two methods of identification were used – one chemical and one biological. Certain functional groups such as an aromatic were identified through the degradation of ILCO and then the genes of the bacteria of interest were sequenced to find similarities to known genes.
It is quite interesting how the expected outcome and observed outcome of this particular experiment differed. The method on this section seemed to place a focus on intending to observe the degradation of aromatics in the Arabian light crude oil, although the results explained that only aliphatic components of the oil were degraded.
I agree that more research should be done before implementing Rhodococcus sp. CMGCZ as an environmental PAH degrader. It seemed as though some of the observed results from this experiment did not match the expected results, which may mean there is more to be considered in regards to PAHs and degraders.
I wonder where there obligate marine bacteria reside mainly. It would be interesting to know their exact locations – such as being coastal or in deep waters.
I would be interesting to know how the abundance of Cycloclasticus either increased, decreased, or stayed the same following the oil spill in the Gulf. This bacterium seems very relevant to this area.
Is it possible that PAHs are more plentiful relative to other carbon energy sources in marine environments so bacterial strains have adapted to utilizing this resource more?
Is 50 micrograms/ mL a fairly common antibiotic dosage? In other words, I’m curious as to why this concentration was chosen.
This paragraph provides another great example of how much overlap there is between organic chemistry lab and microbiology research.
It’s interesting how orf7 in A is read in the opposite direction from every other open reading form.
orf7 was the open reading frame that was read in a different directions from the other orfs. Is it possible that this is an anomaly considering no function or obvious homolog was found?
I think it’s really neat that these genes can be introduced into E. coli as a way to better study the importance of the gene. I’m sure E. coli has been studied so extensively that it’s quite easy to observe differences.
I also found this very interesting! Only 44% similarity in amino acid sequence seems to leave a lot of room for different protein folding.
Is the PAH dioxygenase system a newer evolutionary product seeing as it shares many electron transport proteins with other redox systems?
I’m very interested to read about why they could not find the PCR products for arsenite oxidase genes even though this specific oxidation activity was observed.
It’s pretty interesting how manipulating the oxygen conditions can influence the oxidation and reduction of different elemental forms of As.
I think this is a very cool experiment, tagging specific elemental oxidation states. I did not know this experiment type would allow for different oxidation states to be known.
I wonder if the anaerobic in the dark has a broader temperature range because on environmental adaptation. In dark locations, it could be very cold or extremely hot if near a thermal vent. I would expect light conditions near the surface to be much more consistent in temperature.
I also thought this finding was interesting. It seems to me that at 50 C some sort of denaturation occurs and prevents further activity from occuring.
From this paragraph it seems that the researchers concluded that acetate is utilized as an electron donor as a result of photoheterotrophy.
Acetate was not utilized in the reduction of As (V) because that reaction occurs in the dark and results from chemoheterotrophy. The difference in energy sources results in different molecules being utilized in the respective redox pathways.
I think it’s pretty neat that although acetate itself couldn’t be utilized in the reduction of As (V) its byproduct, CO2, could be so radio labels from acetate are visible in As (V).
You made a very good point. Would the bioremediation of Rhodocuccus for PAH degradation purposes have unintended or potentially harmful side effects on different aspects of the environment? I did not think about that while reading this experiment, but it seems necessary to answer your question before proceeding to introduce degraders into the environment.
I’m curious about how the researchers figured out the composition of the hot spring water in order to mimic the environment. It seems like the chemical composition could possibly shift due to certain environmental effects and not be entirely consistent.
Jesse,
I also thought this part of the materials and methods was interesting. I’m curious if the researches dismissed these two clones because they suspected issues or if there is any possibility that studying these two clones could lead to possible new discoveries.
I know you can do mutagenesis to enhance the compatability for transformation and expression using E. coli. They may have found some problems with all but ArhA1A2.
In paragraph 4 it mentions that multiple clusters can be found within the genome of some sphingomonads. They seem to wanting to find out how these clusters work if the genes aren’t being regulated within the same operon.
I remember that mutagenesis can be done to aid transformation and expression in a host. ArhA1A2 may be easily transformed while the others need to be mutated first to be accepted by E. coli
By paragraph 4, It seems as if the approach is to find out how the clusters come together to work on the same pathway when the genes are not coordinated through the same operons.
Would they be using this knockout to observe it’s effect, or lack thereof, to determine it’s link to the genes that have already been studied?
Could they have also done mutants without ArhA1A2, and also take out ArhR to see if its effects can be seen on the A4 electron transport proteins? The same could be done without the ArhA3A4 genes instead of A1A2 to assess which genes it possibly inhibits or induces.
I know that cosmids are able to utilize the cos site as sticky ends like a phage vector, but the extra phage genes are gone. Transduction happens really easily with this type of vector so maybe that plays a part.
Would this be an important find because all of the genes studied are downstream of ArhA3 and upstream of ORF6? Would this suggest that these genes are all required to function as a cluster and not be located seperately?
It seems as if AG3-69 doesnt grow on acenaphthene or its intermediate 1,8,-NA suggesting that the mutagenesis carried out removed the strains ability to oxygenate acenaphthene.
The marine environment is a huge dump for pollutants, I would assume that by now these organisms probably have many acquired genes for degradation of PAHs and would prove to be an asset to studying mechanism pathways of multiple degrading genes.
Are the bacteria being studied from the family Rhodobacteraceae too? I’m unsure if they are or if they were just pointing out another organism who displays flouranthene degradation.
Would the genes being located close by have anything to do with how many genes they are finding? It seems they have located way more genes than the previous papers.
Are these metabolites produced during degradation of a specific PAH or are they just providing a list of metabolites produced as a whole, rather than for flouranthene degradarion alone?
Could they have also checked for the lipoproteins and glycolipid’s mentioned in paragraph 15? They could have assessed whether they are constitutive or based on response to fluoranthene.
Do we know if B30 can degrade fluoranthene? If the genes which aren’t shared with B30 are important for degradation, could they just enhance the ability of the B30 genes found in P73?
I think a microarray could be a convenient way to assess which of the many genes they have found are produced under an environment containing solely fluoranthene as a carbon and energy source.
What is the significance of this gene being in the toulene/biphenyl family?
When compared to the other studies it seems this one gathered more information with the methods they used. The way this organisms PAH degradation genes were more closely organized seems to have benefitted the results of this paper.
“However because siderophores are released from cells, they are considered public goods and are susceptible to exploitation by non-producing cells”
Vibrios bacterial species utilize cooperative behavior. An environment with limited resources affects cooperative behavior by increasing costs for the producing cell, because resources are released away from growth towards cooperative functions as public goods. Siderophore production qualifies as a cooperative benefit. Many species are siderophore non-producers but have the genes to bind and utilize it. So these “cheaters” benefit from the cooperative actions of others without suffering the cost and energy themselves.
A gene knockout will cause the DNA to mutate in such a way which prevents the expression of a specific gene. Here, Vibro chromosomal mutants are generated by introducing plasmids incorporating the V. Fisheri genes iutA with fhuCDB. Assumingly, these genes enable V. fischeri to prevent the growth of other vibrio species including V. harveyi.
I researched the co-culture competition between V. fischeri and V. harveyi, under iron-depleted conditions. Aerobactin, a bacterial iron chelating agent production allows V. fischeri to competitively exclude V. harveyi— which does not have aerobactin production and uptake genes. In contrast, V. fischeri mutants incapable of aerobactin production lose in competition with V. harveyi.
And introduction of these genes are sufficient to convert V. harveyi into an aerobactin cheater.
Once the microorganisms completed their purpose, would they continue to be benign? If not, would measures be taken to eliminate them?
What is the next step if no naphthalene degrading bacteria can thrive in a highly selective environment? Would alternate bioremediation strategies need to be considered?
Was this step performed in order to confirm that these microbes were NP degrading bacteria, or was it performed in order to further characterize them?
How does this fact translate logistically for biodegradation of oil and pollutants? Would strain N7 be the best and/or most efficient biodegrading bacteria?
When choosing a bacterial strain to use for biodegradation, is emulsification activity or BATH a more important indicator?
How does the knowledge of optimum concentration translate to the practical use of bioremediation?
Is one of these two types of NP-degrading bacteria (gram-negative or gram-positive) more eco-friendly than the other?
Since the gene coding for the most important member of meta-cleavage pathways is located in plasmid, how difficult would it be to bio-engineer non-PAH-degrading bacteria to be PAH-degrading?
Was this strain anticipated to be a good model organism because of the characterization of the catechol meta-pathway operon genes and their neighboring glutathione-S-transferase gene?
Why is identifying the different degrading bacteria strains and their efficiency important? Based on which bacteria is the most efficient, would environmentalists introduce those strains into environments that need more bioremediation than what was naturally taking place?
I looked it up, and the way I understand it is Biolog-GN tests are a way to determine bacterial strains’ ability to grow with different carbon sources. Is this right?
Is the slant medium referenced in this paragraph similar to the slant medium we used in the previous lab?
I don’t remember reading it, and I looked back and didn’t see anything in the Materials and Methods. Is it mentioned in the paper how they determined the fatty acid composition? Would gas chromatography be used?
I read online that GST was a class of molecules best known for detoxification. Does this mean that GST is directly responsible for PAH-degrading bacteria’s ability to be used for bioremediation?
I looked back at paper 1. At 200 ppm naphthalene, the optimal strain had 89.94% naphthalene degradation, and at 100 mg l-1 phenanthrene, this strain had 98.74% phenanthrene degradation.
The fact that they mentioned this strain could degrade phenanthrene “thoroughly” made me wonder if it is possible that other degrading bacteria only partially degrade phenanthrene.
Does this meta-pathway arrangement account for this strain’s efficiency at degrading phenanthrene?
Why would they have not been able to obtain authentic PCR products for arsenite oxidase genes? Were they not able to design a primer suitable to these genes?
For respiratory As(V) reduction, is As(V) the final electron acceptor? What is the electron donor?
This question was answered this week in lab. When this paper was written, the GenBank only had a few sequences to design a primer. The primer designed was probably not suitable for these particular genes.
What does this mean that these clones failed to show significant similarity to anything in the GenBank database? Does this mean that no one has annotated these genes yet?
If the arsenic speciation and acetate concentration were measured by HPLC, was the purpose of radioactively labeling them to make them detectable?
Why did the reduction/oxidation rates diminish each time?
Why was the sulfide condition not tested for acetate assimilation?
The percentage is above the baseline number given with no additions; therefore, assimilation using acetate was stimulated by the addition of As(V) and As(V)+H2. It was not stimulated by the addition of H2,killed. The number for the addition of H2 is too close to the baseline number to give a definite yes or no.
So in the light, As(III) acts as the electron donor (what water would do in aerobic photosynthesis) in anoxygenic photosynthesis, and in the dark, As(V) acts as the terminal electron acceptor in anaerobic respiration (how oxygen would act in aerobic respiration)?
I think you answered this question in lab today, but I cannot remember what you said, and I didn’t write it down. In figure 4, why was there a slight reduction of As(V) when no electron donor was added?
Why is it that this gap in knowledge exists for marine-sourced degraders? Are these harder to isolate and/or sequence?
What determines which intermediate the PAH will be transformed into?
I Googled paired-end sequencing; is it just sequencing the DNA segment from both ends? Why would they choose to sequence the DNA this way?
Are suicide vectors always used when performing recombination, or is this particular to the cre-lox method?
Why would this strain possess genes for chemotaxis, yet be non-motile/not possess genes for flagellar proteins? Why have they not lost the genes for chemotaxis?
Only one pathway is referenced in this paragraph; however, would the strain utilize one of multiple pathways depending on the intermediate?
When ring hydroxylation is labeled “the most difficult catalytic step”, is this meaning it’s the most energetically demanding?
How do PAHs compare to other compounds as a carbon source?
I don’t understand the significance of this gene being of the toluene/biphenyl family?
To further study the functions of the catabolic genes. would they perform more gene knockouts? What else would they do?
Why does the presence of heavy metals inhibit the enzymes? Is it a form of noncompetitive inhibition where the metals are interacting with the enzyme?
Why is information on this topic limited? Has little research been conducted, or has research been conducted and not yielded a lot of information?
So for this experiment, the amount of PHE degraded should decrease as the concentration of Cu(II) is increased because the copper hinders the activity of the degradation enzymes. Also, will they test each initial concentration of PHE with each concentration of Cu(II)?
For the blank control, would PAH-RHD and C230 genes likely not be transcribed at all?
From what I understand, they’re interested in the effect of copper because the PHE needs to be degraded because it is a pollutant; however, heavy metals (like copper) can hinder the activity of the enzymes necessary.
Why would the biodegradation rate of PHE in PAHs-contaminated soils improve in the presence of cadmium and arsenic?
From what I understand, was the only unexpected result that the transcription of PAHs-degrading genes was promoted by high level of Cu(II)?
I googled a different comparison of cluster structure, and in that one, the length of the arrows corresponded with the length of the ORFs and the direction corresponded with the direction in which it was read.
Are the PAH-degradation genes frequently dispersed because they’re often acquired through horizontal gene transfer?
The way I’m understanding this paragraph, degradation enzymes have been located and described for a fluorene-degrader; however, it’s gram-positive. Fluorene-oxidizing enzymes have been located and described for gram-negative degrader; however, the hosts doesn’t grow when fluorene is the sole carbon/energy source. This paper will be the first that locates and describes fluorene-degrading enzymes for gram-negative degrader that can grow when fluorene is the sole carbon/energy source.
So for this overexpression experiment, the dioxygenase genes (flnA1-flnA2) were cloned into the overexpression plasmids from paragraph 6, allowed to grow, and then the products from that were extracted and measured using GC as in paragraph 9?
A follow-up to my last question: is the SDS-PAGE also a step in this experiment? Do they perform SDS-PAGE before GC? I get a little confused with the methods not being in order of the experiments.
So if we were to do expression cloning for our project, how would we test if the protein was made? Would we be able to do GC or SDS-PAGE?
The differences in expected and observed sizes mentioned here – are these significant? I can’t tell from the paragraph if this is important.
Should we focus of substrate range? Because I found this paragraph very confusing.
They performed RT-PCR of flnA1 and flnA2 using cloning vectors to see if they were transcribed more in strain LB126 when grown on fluoranthene and not when grown on glucose. This was the case. They then used expression vectors to see the enzymatic activity of these same genes.
Phenanthrene is one of the preferred substrates; however, it cannot be used as a sole source of carbon and energy by strain LB126. Could this be because, like you mentioned in one of your comments in the introduction, the initial enzyme has the relaxed substrate specificity to accommodate phenanthrene, but the later enzymes in the pathway do not?
Looking at the product concentrations in Table 3, it seems as though fluorine and phenanthrene were drastically the preferred substrates.
This paper is looking at the degradation of carbofuran even though several metabolites cause effects on the reproductive system in female rats. Is this because biodegradation is the only way to eliminate carbofuran even if there are negative repercussions?
Sphingomonad strains seem to appear in papers for the degradation of many different substrates. Is this because Sphingomonas is just more capable (than other bacteria) of degrading a wide variety or is it because this is just what is isolated when researchers collect samples?
So for this experiment they performed random mutagenesis on the strain KN65.2 to look for the loss of function (degradation of Carbofuran)?
The gene that when interrupted resulted in the loss of Carbofuran degradation was identified, and then sequenced to design primers in order to perform RT-PCR to confirm function?
I would think this result would be a surprise. All of the degradative pathways we’ve looked at so far in these papers have been inducible if I’m remembering right. Usually glucose would be the preferred C source and then the alternate pathway would be induced when glucose was used up.
The mutants for the most part exhibited reduced or slower mineralization rather than the abolition of mineralization. Is this because the pathway was shown to be constitutive?
So they selected mutants and assigned them to five groups based on whether how they grew, degraded, and/or mineralized carbofuran; they then selected ten of those mutants and verified whether carbofuran phenol (what they think is the first metabolite of degradation) is detected during the process?
How rare is this deviation from the expected regulation of catabolic genes?
So this paragraph is saying that although they used plasposon mutagenesis to identify genes responsible for carbofuran degradation, they had to be careful assigning loss of phenotype to specific genes because the loss of phenotype could be a result of polar effects instead?
Group I: C1 metabolism genes affected -> C1 metabolism is predicted to be related to conversion of methylamine -> hydrolytic release of methylamine moiety is first step in carbofuran-degradation
So for group I, the delayed/decreased growth rate on carbofuran is explained by C1 metabolism genes being affected, which in turn affects the first step of degradation, or am I misunderstanding?
Does the ONR7a medium select for NP degrading bacteria more efficiently than the MSM we used in lab?
I’m very interested to see specifically how they collected the bacteria and recreated an environment similar to their habitat. If the bacteria proves to be successful, how long will it take before they can actually start using this bacteria in the wild?
I figured that the oil eventually floats to the bottom of the sea floor, but I always wondered how it impacted the organisms down there since it’s already a pretty extreme environment. If bacteria is used for bioremediation here, would the goal be to remove all of the oil or just decrease the concentration?
These methods are super dense, but I love that I can recognize the primers and semi-understand the processes. Why did they use “near complete 16S rRNA gene sequences,” I feel like it would make more sense to use fully complete sequences?
What is the special characterization of ONR7a agar plates and why was it selected?
I’m confused. I thought only Eukaryotes have mitochondrial respiration and Bacteria has respiration through cell membrane. If Naphthalene hinders mitochondrial respiration, it would not do anything for the bacteria, right?
If we were to use one of the common naphthalene degrading bacteria against a PAH compound that has not been well characterized and its not able to compete with the bacteria that’s polluting the water and ecosystem, will the selective pressure make the bacteria that we are trying to fight worse and even harder to get rid of?
Since the N1 and N2 strains has the most growth, does this mean those are the two stains that are causing the most pollution in the Persian Gulf? If so, would bioremediation of these pollutants significantly reduce the pollution in the water?
I was looking at the phylogenetic tree tables, and I was wondering if the originator of all the stains was targeted with the bioremediation approach would all the strains that followed be eliminated as well?
Since the different genera produce biosurfactant and bioemulsifiers that can uptake and dissolve the component of the crude oil that is contaminating the Persian Gulf already, is there a need to anything else? It seems as if the ecosystem is fixing itself.
Would it be better to have more N1 and N7 strains in the Persian Gulf so that more of the oil contamination can be dissolved?
It seems like they should focus on the obligate marine bacteria in coastal marine environment versus terrestrial habitats since this bacteria has only partially been studied. They are studying something that has already been extensively characterized.
I don’t feel like the author really told us the importance of their study as opposed to all the other studies on this topic. What will this paper tell us that the others didn’t ?
What makes ampicillin and kanamycin the best antibiotics to add to the media in order to select only the bacteria with recombinant genes?
Why was the library replica plated onto LB agar plates with kanamycin and not ampicillin also?
If the large subunits do not have small subunits to pair with, what will happen?
Since they did not have any small subunits to make their dendrograms, did they just use the previously published amino acid sequences based of the large subunits found in their experiment?
Is there any benefit to having gene clusters responsible for PAH degradation on the plasmids rather than the chromosomes and vice versa?
What does it mean for a gene product to fall outside the major cluster?
Why does the rate of oxidation and reduction of the samples get slower each time the light and dark regimen is shifted?
Is the reason why the killed controls neither oxidize or reduce because of their proteins being denatured by heat?
How did they come to the conclusion that As (III) was oxidized due to anoxygenic photosynthesis? They should not have been able to conclude this just from this one figure. Couldn’t the oxidation be due to chemolithtrophy since it can occur in the light or darkness?
So does this mean that As(III) will continue to reduce due to the microbes losing their ability to oxidize and not due to the microbes being incubated in the dark?
Why do the 16s genes have to go through PCR amplification in order to be identified? If all PCR amplification does is make more copies of the gene, why can’t the single gene be identified?
Why did the samples include both contaminated sea water and marine sediment ? If the sea water and the sediment was collected at the same location wouldn’t it have the same type of bacteria? Why the need for both?
Why were they unable to obtain authentic PCR products for the gene even though they observed aerobic oxidation activity? Will they elaborate more later on why they were unable to obtain PCR products?
Since the Ectothiorhodospira bacteria is dominant in the biofilm, does this mean this bacteria has the best resistance to arsenic opposed to other bacteria ?
Does arsenic support anaerobic respiration since the medium they used was oxygen free ?
What is the signicance of the shaking of the incubated tubes ? Does this have something to do with the electrons being donated ?
What does PAHs stand for? Do these compounds referred to in this paragraph still pollute the environment badly in the U.S. because of sewage waste water and home sewage?
How much cleanup has bioremediation done since the oil pollution during the Persian Gulf War? How affective is PAH bioremediation really since there is already so much damage?
When did researchers realize that cyclic aromatic compounds are harmful to mammals? How long has this fact been known or in literature?
I know that in lab we pour agar. Do you turn solid agar into liquid for GC purposes? How would you turn solid agar into a liquid to extract the different compounds mixed in it?
What is starch hydrolysis? What would be the significance of starch hydrolysis in this experiment?
Why were 18 out of 54 strains isolated? What happened to the other strains that made them not have an adequate growth rate? Could some of these strains be cross-contaminated?
What is a shaker incubator? Why wasn’t the microbial growth and naphthalene biodegradation monitored every day instead of just 15 days after? What made the experimenters choose 15 days of growth and then monitoring?
It would be interesting to read about why researchers think that Gram-positive bacteria might play a role in naphthalene degradation in the highly variable environment of oil-contaminated sediments. What brought them to these conclusions?
Are there any other regions that have a high diversity of naphthalene-degrading bacteria besides the Persian Gulf?
In this study, did the researchers choose to study Gram-negative bacteria instead of Gram-positive or was that just a coincidence?
How do researchers know which bacteria types are best for bioremediation? What tests and check-marks were used to come upon such assumptions?
Was phenanthrene-degrading bacteria chosen to study because is an aromatic compound? Are we going to discover/read if naphthalene-degrading bacteria are better or worse than phenanthrene-degrading bacteria?
What were the identifications and characteristics of distinct colony morphology? What were the researchers looking for?
Why did the amplification have to go through 29 cycles of denaturation? Was is the “extension” process?
Was there only one isolate that was found that would be able to degrade phenanthrene? This is different from the last article because those researchers found several strains of bacteria capable of degrading naphthalene. Does this possibly mean that phenanthrene is harder to degrade because there was only one isolate found?
There are quite a few aromatic compounds that Strain ZX4 can use for carbon and energy sources.
There are quite a few aromatic compounds that Strain ZX4 is able to use as carbon and energy sources.
Out of the 95 carbon sources, what were “wrong” with 40 of the sources to where Strain ZX4 could not oxidize them?
I am interested to know what was the two percent difference between strain ZX4 and strain UT26, since they had a 98% similarity.
In this paragraph, it says that strain ZX4 could be a potential strain used for bioremediation. What other types of tests and research would be need to know if this strain could be used for bioremediation rather than just having the potential to be used?
Should the initial step involving meta-cleavage of catechol have been catalyzed by PhnI, but it was done by PhnH instead? I am a little confused on the first statement.
What is the salicylate pathway?
Why did the researchers not focus on the cycling of arsenic under oxic conditions too other than just focusing on anoxic conditions?
Why were the researchers unable to obtain authentic PCR products for arsenide oxidase genes? Will this be discussed more in the study?
Why were the researchers unable to obtain authentic PCR products for arsenite oxidase genes? Will this be discussed later in the paper?
Why were the slurries stored in the dark before being used in experiments?
Why were only groups containing two or more clones sequenced?
Why did the rates not change after constant shaking of the samples? I would think that after they shook the samples to increase oxygen exchange then the rates would favor oxic conditions.
What is a hypersaline environment?
So with increasing ever 100h of incubation, does the rate of As(V) reduction further diminish every time?
Is acetate a key terminal substrate in aerobic process or is it just one in anaerobic processes?
When this paragraph says, ” We recently proposed that the respiratory arsenate reductase of strain PHS-1, and presumably the very similar sequence found in the biofilms, functions in vivo as a de facto arsenite oxidase”, what does “in vivo as a defacto arsenite oxidase” mean?
Will the researchers continue to search out this unexpected result? Will they research out about why there was an observed aerobic As(III) oxidation carried out by a novel mechanism? Will they see what this “novel mechanism” is?
About how long does the process of bioremediation take in order to efficiently eliminate all of the environmental pollutants in the area?
I am curious to know the effect of more complex PAH’s on certain bacteria. Would they be able to efficiently eliminate environmental pollutants or would the by-products be malignant to the environment and people?
Why is it necessary to transport the seawater on ice when the samples were gathered from just 15cm below the surface? I feel as though the ice would bring the temperature above normal conditions therefore having effects on the organisms that live within it.
How did they know what temperatures and times to hold the PCR at during the 35 cycles?
Why does the increment pattern cease at 400ppm concentration?
I am wondering the same thing. Is the GC-FID method some sort of gas chromatography method?
What factors play a role in the reason for the high diversity of naphthalene-degrading bacteria in the Persian Gulf besides the pollutant. If this is the only reason, could the same experiment be done in a different part of the world that has a similar pollutant?
I am curious to know what the optimum concentration for naphthalene-degradation would be for isolates found in different areas besides this specific area in the Persian Gulf.
What characteristics of the strain made the researchers anticipate that it would be a good model organism for their experiment?
What do they mean when they say the catechol 2,3-dioxygenase mirrors the taxonomic grouping of the host bacteria? Does this mean that it possesses similar characteristics of the host bacteria?
What is the purpose of amplifying the 16S rDNA? I see that you are basically breaking the strain down, building it back up again, and I think making it longer? But what purpose does that serve?
Why use the supernatant for enzyme assay rather than disposing of the supernatant and using a different enzyme? Does this result in different enzymes being assayed?
What are BHY plates? And what are some benefits of using these types of plates over others (i.e. R2A, MSM, etc.)?
I found that the Sphingomonas paucimobilis strain is commonly found in hospital equipment such as vents and can cause disease. I think it is interesting that a strain with these characteristics also possesses positive characteristics in the sense that it is able to degrade phenanthrene which is found in cigarette smoke.
Is the pH range for this strain considered a wide range or no? I would assume that a 2.5 difference would be considered a wide range. This should allow for a greater variety of conditions that this strain is able to grow and survive in compared to other strains.
I think its extremely interesting that this ZX4 strain offers an environmentally friendly alternative to creating indigo instead of having to harvest it from plants. Another positive capability of this strain is its ability to be used in bioremediation. Two environmentally friendly uses.
How would the researchers go about obtaining information on the structure of the meta-pathway?
I looked up what xenobiotic compounds were and have concluded that the ability for the ZX4 strain to degrade these compounds is yet another environmentally friendly characteristic that this strain possess. What are some environments that xenobiotic compounds can be found in? How can this strain be introduced in those environments in order to rid the environments of those pollutants and how long will it take for them to be completely eliminated?
Did the researchers decide to focus on examination of the cycling of arsenic under anoxic conditions because of how the coculture responded in the manipulated laboratory conditions?
What makes the biofilms used in this experiment a good model system for this oxidation/reduction reaction?
I was wondering the same thing, but also what made the green-colored springs and small ponds so rare compared to the red springs?
How did they keep track of the tubes that were shaken before they were inoculated and the ones that were not? Is there any specific reason for shaken some of them and not shaking others. I am curious to see the results of the tubes that were not shaken and and how they may differ from the tubes that were.
I believe they did not include single clones in the sequence analysis because it would not be financially worth it.
So, since the rates did not change can it be said that the conditions of the environment, whether its oxic or anoxic, does not play a major role?
How could they test to determine what the functional gene is since it is not aoxB even though it shares similarities?
How could the authors know for sure whether the capacity for aerobic Arsenite oxidation is due to the shallowness of the pond or if it is another reason?
What are some ways that the researchers could determine which biofilm microbes that were associated with the arrA amplicons?
What is a novel mechanism?
Do all regions have unique PAH-degradation patterns or is only the Nakheel region unique from the others? I would assume that if each region has a unique pattern, then removing PAH’s completely would become an extremely difficult task.
They mention how the concentration of the PAH is important for removal. Are the co-substrates like yeast used for this purpose, or how do they go about lowering the concentration of the PAH?
I am a bit confused by this paragraph as to why they are testing the sediment samples ability to withstand the stress. What does it mean about the sample if it is not able to do so?
What is the advantage of constructing the phylogenetic trees? Could they use information from them to determine similar degradation methods that have already been determined, or would this not be possible with the unique PAH samples they have found?
If bio-emulsifiers “act like surfactants” and the water solubility of LMW PAHs is improved when surfactant-production is present, could these bio-emulsifiers ever be modified to be useful for both LMW and HMW PAHs?
I know they mention that there is no significant difference among the 3 sites, but table 1 shows that there is a slight difference in temperature at all 3 sites. Does this have any impact on the concentrations of the different nutrients found at each site?
I think it is pretty interesting how the table shows that none of the isolates showed ‘excellent growth’ when it came to a PAH substrate. They were all ‘good’ or ‘poor.’ Could that mean there are much better strains to use for bioremediation?
If all of the isolates were either at 100% similarity or very close, then why would they not look at different sequences aside from the partial ones they did? I guess I am wondering how they are able to draw any unique conclusions here if all the results are almost identical.
My question is a bit related to both papers, so if hydrocarbon contaminants can be toxic in the waters, wouldn’t evolution have created better mechanisms within microorganisms for treating them? Or has not enough time passed yet because most of these pollutants are more recent and man-made?
Does the temperature of the environment have any effect on the hydrocarbons that are present?
It says that the cores were stored for subsequent use within 2 weeks. Does the storage time have any effect on how the following experiments on the core go? Or would the results be the same as one day later or two weeks later?
16s rRNA sequencing has been in both of the papers we have read. Is is just a common thing in general for sequencing or just for use in studying types of microorganisms/PAHs in these types of marine environments?
All of the genera in this paper are different from the last paper. I just find it kind of interesting how there seems to be a lot of species with potential of being PAH-degraders but it is still a huge problem in the environment.
I am a bit confused on obligate/non-obligate. I know what those mean in general but does it mean here that the obligate PAHs only responsibility is to degrade them while non-obligates have other uses in addition to degrading?
How did they select these PAHs as the ones to test the incubations on?
What exactly is a singleton sequence and why did it mean that those two weren’t further analyzed? I tried to google it but all the answers were a bit confusing.
They say that Se-reducing bacteria is ubiquitous so is the high concentrations of toxic forms of selenium natural or caused by something?
I am a bit confused because the previous paragraph says only two reductase genes have been characterized but this paragraph talks about a separate one, or am I reading this incorrectly?
How do putative knockout mutants differ from other mutants aside from the fact that they didn’t grow on the LB agar?
I am a bit confused here. So they are trying to screen the E. cloacae library and they need a strain that doesn’t have selenate reductase activity? Why is that?
So were FM-1,2 and 3 all just separate isolates of the mutated SLD1a-1?
So the pECL1e clone was chosen for further characterization because t he pLAFR3 showed no ability to reduce SE(VI)?
So just to clarify, the fnr gene was inserted into the S17-1 and that is responsible for oxygen-sensing and therefore responsible for reducing Se(VI)?
So the Se reducing ability is not related to presence of oxygen? The presence of oxygen only applies to the fnr gene?
After reading the introductory paragraph, it’s clear that individuals exposed to PAHs mainly suffer from symptoms relating to the respiratory system. My question would be which type of exposure yields worse symptoms, dermal uptake or respiratory uptake? And what are the various types of preventive measures that have been put in place to avoid exposure to PAHs?
The article briefly mentions how both LMW and HMW microorganisms are used as a means of bioremediation against PAHs. However, does one prove to be more efficient than the other? Is the reason LMW microorganisms are more commonly used in studies because they’re more easily accessible or because they’re the better option of the two?
In paragraph 2.3, the procedures detail how they let colonies grow on MSM plates for approximately two weeks before the isolating and sequencing the cultured colonies. However, in paragraph 2.4 the procedures mention how they only let the naphthalene degrading bacterium grow on MSM plates for 8 days. Why weren’t the plates given 2 weeks to grow? Did they not need as much time to grow?
What difference did using the YMSM plates use for the experiment? What was the advantage of using YMSM over regular MSM plates?
For the MSM plates, why were the percentage degraded for Nap and Phe so close compared to the Fla? Are the chemical structures for Nap and Phe somewhat similar? And if that’s the case, shouldn’t they also have similar percentages degraded when placed in the YMSM plates?
Given that the degradation percentage of crude oil was so low, would it make sense to search for an alternate degrading bacteria? Or are these results typical to what you’d expect when attempting to degrade crude oil via bioremediation over such a short period of time?
It’s definitely interesting that CMGCZ was better able to degrade Fla more efficiently than a structure like Napthalene. Especially considering Napthalene is only a two ring PAH while Fla has four rings.
It sounds like the addition of YE in a medium didn’t lead to a significant change of results. Why was YE chosen to be used in the first place?
Given that we’ve already read about the bacteria Fla, I wonder which one (Fla or Cycloclasticus) is a more effective means of bioremediation
I accidentally compared Fla and Cycloclasticus.. when Fla is a PAH and Cycloclasticus is a degrading bacterium. So my original post should’ve asked if Cycloclasticus is a better bacterium at degrading Fla than the original bacterium we studied, Rhodocuccus.
Of the PAHs being studied, I wonder which ones are of a low molecular weight and which are of a high molecular weight. Which will be easier for the Cycloclasticus strain to degrade? Does it have anything to do with the marine environment?
The LB agar plates are the same used from the first experiment we read about for Blog Summary 1. To me it seems to be a standard growth plate used in labs everywhere.
After reading the introduction of the paper, I thought that the mission of the experiment was similar to the first blog we read; therefore leading to similar experimental methods. However, the further I read into this paper, the more complex this experiment seems. I’m interested to see how the results vary between the two experiments.
I wonder what the specific location of the 10.5-kb SauAI fragment was. And if it was in identical locations in both the pH1a and pH1b constructs. In Experimental Designs with Kroetz, we learned that the location of a specific sequence was key in determining its characteristics/influence on the DNA.
Does this paragraph hint at the idea that there’s a specific sequence of DNA that gives microorganisms the ability to degrade PAHs? It would seem to be the case considered there were 7 cases of PAH-degrading constructs found with similar if not the same sequence.
Why were the chemical structures from Fig 4.4 converted to a different form? What was the purpose, especially considering the form they were transformed to were stated to be more structurally unstable.
So does this hint at the chance that having a shared dioxygenase system is a contributing factor of the increased ability of a strain to degrade a PAH?
This may have went over my head but what is an ISP and what processes does it typically carryout?
Does the ability to degrade PAHs go up as more dioxygenase subunit genes are found in a strain? That’s what this paragraph suggests, yet you would think that the attachment of additional DNA would have the opposite effect.
I wonder if a bacteria’s ability to metabolize substances that are normally considered toxic to organisms means that its categorized as an extremophile. This would also make sense because the paper discusses how some bacteria with this ability live in extreme environments such as hot springs.
I wonder if the inability to obtain PCR products for arsenite genes has anything to do with the specimen being collected from an extreme condition (i.e. a hotspring). Why weren’t samples from somewhere else used and would they have likely yielded the same results? Interested to finding out what the next step in their experimentation was.
I honestly was wondering the same thing Justin. I wonder if the details given about the location were significant in a way that isn’t obvious to a reader. Because to me, the details seemed pretty generic.
Why was there no type of pattern in the intervals that the samples were taken? It seems odd to me that they retrieved 2 samples in 2008 only 2 months apart and waited 6 months to retrieve another one in 2009.
It’s interesting reading about how exactly the researchers set up and prepared their experiment. It seems as if they’re was plenty of room for human error. Which may explain the inability to produce authentic PCR products for the arsenite oxidase gene later on in the experiment. I wonder if there is any correlation.
I wonder if there are any major similarities between the bacteria strain compared to the archaean strains. Because to me it seems like the bacterial strain would have to have some of the same properties in order to maintain survival in an environment typically known to be home to extremophiles (aka archaea).
It’s cool seeing how the slurries were able to go back and forth between oxidizing and reducing arsenic based on their environmental conditions. I wonder if the amount of exposure to either light or darkness affected the speed of oxidation/reduction.
This paragraph answers my question that I wrote under paragraph 5. I’m curious as to why the temperature range for reduction is so much bigger than that of oxidation.
How can it be determined that acetate was used as an electron donor when in the paragraph above it’s stated that there wasn’t any observation of As(V) reduction with acetate?
Would the next step be to try to make new and more specific primers in order to better detect arsenite oxidase genes?
This paragraph is important if the reader has no background knowledge of what PAH is or the dangers of it to the environment. It not only breaks down the toxicity of PAH, but also discusses why the chemical is so hard to remove.
This paragraph highlights the overall purpose/goal of the experiment. In the previous paragraph, it is noted that there are bacterial strains capable of surviving in the toxic environment of the PAH stricken Arabian Gulf. The goal of this study is to isolate the bacterial strains growing in the PAH environment and further study their functionalities. A better understanding of these bacteria would help future bioremediation efforts across the world.
I noticed that the researchers used 16S rRNA for constructuing the evolutionary relationships of these bacteria. My question for this portion is; is sequencing the 16S rRNA more effective than the 23S and the 5S rRNA segments?
What makes the temperature of a particular sample site important? Was this to ensure that the sample area could sustain the PAH degrading microorganisms or is it something more technical?
Does the fact that Isolate LC having 100% similarity to P. aeruginosa imply that it is definitely the same species?
What is the reasoning behind the difference in degradation abilities of the same organism on different PAH compounds? Are the compounds so unlike one another that certain bacteria will specialize into a particular PAH compound, rather than PAH’s as a whole?
The PAH isolating bacteria isolated from soil on the surface would have little to no effect on oil degradation at such low depths, could this class of organism be understudied due to the expenses involved in isolating it?
Since these areas are ridden with hydrothermal plumes, could one expect to also search and isolate archaea PAH degrading organisms, as their extremophilic nature would make them a dominating organism in the ecosystem?
Could removing cores from their pressured environment have changed the ecosystem too drastically for the organisms to preform at the same compacity?
Would an PAH degrading archaeon that could sustain higher temperatures preform better the closer one got to the hydrothermal center? Could a selection of two microorganisms capable of degrading PAH, one being a dominant PAH degrading bacteria and another being a more niche archaeon work together since they would occupy separate spaces along the seafloor.
Could Cycloclasticus have an impact on other deep sea enviroments with abundant sediment surface hydrocarbons? Would Cycloclasticus preform better if it was deposited alongside Halomonas, Thalassospira, and Lutibacterium in a new enviroment?
Is this “rusty-yellowish” coloration able to be observed at the site of extraction? It would be interesting to compare the coloration and presence of oxidized intermediates in the original location and the studied organisms.
The prosthetic heme group acts as an Oxygen binding site in hemoglobin of animal cells. Why would an organism utilizing anerobic metabolism of Se require the capture of Oxygen?
Will the selenium metabolic mechanism always produce a similar mineral product, or will a selenium metabolic mechanism from a different organism produce a different mineral product?
Genome sequencing was clearly important in order to identify the presence of known selenium metabolic genes, but would a transcriptomic/proteomic approach be useful in this experiment when searching if an organism contains rna/enzymes capable of metabolizing selenium?
What would be the proper procedural steps to take if the organisms were capable of selenium metabolism, but none of the three strains contained the gene that they were searching for?
When looking at the graph, it is notable that pLAFR3 expresses a relatively constant Se (VI) Reduction, whereas the reduction rate of pECL1e gradually decreases over time.
This experiment was preformed to confirm that the fnr gene is essential for the reduction of Se (VI) to Se(0) in E. cloacae SLD1a-1. The results from this experiment have shown the importance of the fnr gene.
What causes this nirate inhibition on selenate reductase activity. What can be done to further study this complication?
The FNR gene is essential in the detection of Oxygen concentrations, but is also essential to initiation the degradation of Se(VI). Without FNR, no Se(0) precipitation will occur.
If there was a way to mutate FNR to make it sense another molecule OTHER than oxygen (i.e. mutating the gene so that it detected the presence of nitrogen), could you make a pathway that was dependent on the presence of another gas? Are there other similar pathways in nature that are dependent on such gas concentrations.
Apologizing for all the questions in advance, this is just a fascinating concept.
Many questions arose when I first started reading this article in Paragraph 2. For starters, it states that a main concern to Antarctica’s natural region is being damaged by the increase in tourist and other people visiting the continent. When I googled how many people are annually visiting Antarctica I was shocked by the numbers. It states that when Antarctica first allowed for people to visit in the 1950’s only a couple hundred a year visited. Recently, more than 56,000 tourists visited Antarctica during the 2018-2019 season. That is a huge increase in only 60 years for a continent that is mainly an ice wasteland. I can completely see why native Antarctician’s are becoming more afraid of their climate due to the increase in visitors. My guess is that more people will result in warmer areas and increasing the use of various fossil fuels to bring people on and off the continent.
We probably can all agree that various Oil spills and burning of fossil fuels has in fact caused the Earth’s atmosphere to become warmer and also resulting in Antarctica losing areas of ice each year. [ Oil contamination can generate detrimental changes in soil properties, including modifications in maximum surface temperature, pH, and carbon and nitrogen levels (Aislabie et al., 2004).] From this quote in the article, I was shocked to see so many different types of Proteobacteria being affected from oil contamination and my overall response to this is, well that cannot be good, and its not. From my understanding of paragraph 2, Antarctica are in real trouble when it comes to oil and other fossil fuel contaminations and if it is not improved, many of the bacteria essential for Antarctica’s nature could no longer exist and the entire world will feel its effects.
[53 cultures that metabolized phenanthrene were selected, and the concentration of PAH quantified as described below. The three highest metabolizing strains were then selected for further studies.]
It is very interesting to me to see how these scientists are creating 53 different cultures that will ultimately metabolize phenanthrene and their PAH concentration and pick upon the nest three with the highest metabolizing rate. I believe out of the 53 some might even be rather similar, and if you have multiple that are very similar, it must be quite challenging to select to best options to experiment on?
[Briefly, non-polar compounds were extracted from culture media using two volumes of hexane and vigorous mixing for 60s]
From the Phenanthrene quantification section in this article, i wondered why it was so important to use two volumes of hexane and vigorous mixing for 60s in order to extract the non-polar compounds. So when I browsed the internet trying to find an answer to my question, I quickly realized that the main reason behind it is to just clarify the non-polar compound you are trying to extract, while it also helps purify the substances being extracted. This is also why the procedure is repeated for a total of three times in order to make sure the non-polar compounds were fully and properly extracted.
The main part of paragraph 3 that stood out to me was the similarity in how the glucose in both the unexposed and the diesel-fuel exposed were realitivly the same. On another note, the other 2 aspects of the graph showed increases in CFU for the control group as well as the phenanthrene. To me this shows that an increase in phenanthrene is present in the diesel fuel sample.
[Analysis of optimal growth temperature revealed that all isolates exhibit maximum growth rate at 28°C (Figure 2C), indicating that these strains are psychrotolerant rather than psychrophylic, a behavior we have seen in previously isolated Antarctic strains]
The main question that I derived from paragraph 4 was that why were all of the samples experience maximum growth at 28 degrees Celsius? It is just strange to me to think that the variety of different samples all hit their maximum growth rate at the same temperature rate. As stated in the paragraph as well, this means that the strains are psychrotolerant from our previous examples from the Antarctica stains.
One thing that stood out to me in this paragraph was the fact that Antarctica is a certain region where bringing over foreign organisms is forbidden in the continent. Like the article continues to say, this increases the use of native bacteria samples and also will help narrow down decontamination techniques and strategies. This is something I never knew was true and can fully understand and see where Antarctica are coming from in the fact that they don’t want to cause anymore unwanted bacteria to grow.
To respond to your question Rachel, I agree with you on the point that they were just trying to achieve their main goal, but I also believe because there are so many different varieties of bacteria in the area they are testing, it is more likely that they will not have a completely isolated degrader.
After reading this paragraph, the statement about the variety of competitive strategies intrigued me the most. From raid culture of limiting resources, to contact-dependent delivery of other toxic, can ultimately effect competitive cells to change.
I was confused on the part when the writer explains how species can produce siderophores that will have a competitive advantage to other cells, but what eventually caught my eye was how when these cells are released they are public goods and are exploited by non-producing cells. I found this very interesting and believe to be very important for the upcoming experiment.
There is so much going on in this paragraph it is difficult to grasp what the author is trying to present. Other than all the equipment and type of bacteria’s/ antibiotics. What was the point of so many different samples that had to be added? What was the main advantage for doing so?
I found it interesting that the scientists did not filter the remaining mutant just for simplification purposes, but also how come ploymyxin B was added to prevent any more growth. I’m curious to understand how this actually works.
[Finally, the presence of V. fischeri ES114 culture fluids prevented the growth but did not kill V. harveyi. While below the level of detection by OD600 measurements, a small increase in V. harveyi cell density could be detected by counting colony forming units (CFUs) (Fig. S2D)]
I found this sentence to be very interesting to me and caused me to form a question in my head. I was wondering why does this culture prevent the growth, but still does not kill the V. harveyi. Usually I would assume that if a process or anything is stopped growing by an outside force, majority of the time that product will eventually die off.
[It was curious that growth inhibition of V. harveyi occurred only when culture fluids were obtained from V. fischeri ES114 grown in minimal marine medium but not in rich medium (Fig. S2B). ]
I found this interesting as well, to the point that the inhibitions were only obtained from V. fischeri ES114 when grown in medium but not in rich. I believe this is because the medium was the minimal required for the product to actually grow, and too much medium like in rich, the product might not like and abundance of medium and could harm or damage the process.
[Alternatively, the genes encoding IutA and FhuCDB could be acquired by horizontal gene transfer. Horizontal transfer is known to have distributed aerobactin genes across multiple vibrio phylogenetic lineages ]
This was a statement that caught my eye and I thought was quite interesting. I did not realize that horizontal transfer is known to distribute aerobactin genes across multiple lineages. I believe that this is actually necessary for the given experiment, and without them, it could be hard to encode these genomes, like with siderophores for an example.
[It could be beneficial to repress iron uptake under oxidative stress because ferrous (Fe2+) iron reacts with hydrogen peroxide in the Fenton reaction to generate harmful hydroxyl radicals that damage DNA (Imlay et al., 1988).]
Another statement in this section that I found interesting was the fact that more iron uptake under oxidative stress can be better beneficial. This is very interesting in the fact that they could have been using this technique for a long time in the experiment, and also helps understand how some things are changed and processed in this experiment, that could eventually harm and damage the DNA they are sequencing.
The fact that nitrogen recycling is important for marine life and having a nitrate reservoir, is something that I did not know and found very intriguing. It is interesting to see how nitrification can play a role in promoting marine and nitrogen loss.
[Since then, similar sequences of crenarchaeal AMO gene subunit A (amoA) (69–99% amino acid homology) have been detected in various marine water columns and sediments, including the Black Sea (20)]
I wondered why are these sequences so similar? And also the black sea caught my eye as well and also wondered if this body of water had some extra resources containing nitrogen than other seas. Since there was in fact various water columns and sediments in the black sea.
I was interested in what the CTD looked like and how it performed, so after looking at the link Dr. Ni Chadhain, I had a more understanding of how the equipment worked. Overall, the picture helped me better understand what was going on in this paragraph, because I gained an idea of how the machine works.
I was interested in how a FISH actually worked and why the CARD-FISH was better to use than the other. After reading some more information about it and following the link attached above, I can see why they used this system because it does indeed work well will samples that have low abundance. The CARD-FISH is shown overall as an improvement when compared with the normal FISH mainly in aquatic environments.
In figure 2, after observing the charts, I had a hard understanding of them but after todays class where we had an example of this figure, I have a better understanding. Although Figure 2d was some what confusing but can be seen to show the relationship between a,b, and c.
I feel like this figure was easier to understand than the Figure 2, mainly because each color has a different label. From this, I believe that NO2- is probably the best source of vertical distribution in water, since its lines are so close to the edge of the chart. It is also good to know that O2 levels are very high near the surface, and of course very low as the water level drops.
From this graph it was similar to the others that we discussed. Since the oxygen gets lower as the water depth drops, this can help apply the theory that sulfur reducing bacteria will successfully live in. Nitrogen can also be shown in the water levels as well.
Its good to know that this was a vertical distribution of the amoA expression with the crenarchaea. The crenarchaea levels seem to be the greatest as the water level drops. Also this can be seen in the graph in figure 2 where it is better represented than betaAOB and gammaAOB.
This part of the introduction provides background information to the reader. It explains what PAHs are and how they’re harmful as well as a summary of how and which microorganisms break them down. However, the authors caveat by saying that there is much to PAH degradation that we still don’t understand, especially with regard to high MW variants.
It seems according to this paragraph that while we’re capable of sequencing the genomes of PAH-degrading bacteria, without a better understanding of the metabolic pathways that actually serve in breaking down PAHs, the knowledge of these bacterias’ genomes is of limited use to us in understanding the breakdown of PAHs.
I am somewhat curious as to how the sediment for C. indicus and C. baekdonensis were extracted from the ocean, as well as what depth they came from (even though it’s not exactly relevant to the article). I also tried looking up Ausubel et al. DNA extraction, but most of what I found were similar citations to it.
I looked more into GC-MS and it was pretty interesting. Apparently the SCAN mode, if I looked at the description correctly, is useful in determining unknown compounds and scanning ranges of mass fragments. I’m not entirely sure what it’s being used to determine in this paper, but I read that GC-MS can be used to determine metabolic activity, which I suppose could be useful in trying to isolate a pathway for PAH degradation.
So to see if I understand this figure correctly, this figure (along with paragraph 11) is showing the predicted phylogenetic tree containing P73 and its 5 plasmids present, and that the trees are based off comparing the protein sequences of the parA gene (for figure 2A) and the rep gene (for figure 2B) with other available genomes, and that the closest related bacterial species to those plasmids is where they might have originated from?
Interesting that the P73T strain has the genes for flagella assembly, motility, and chemotaxis and yet phenotypically demonstrates none of those traits. Does simply having those genes assist in increase bioavailability of PAHs, or is P73T perhaps relatively closely related to a strain that is both motile and capable of breaking down PAHs, and this strain for whatever reason lost the ability to assemble flagella or become motile?
So the way GC-MS can be used to determine metabolic pathways is that it can detect potential products and byproducts of degradation, like it did here for fluoranthene degradation?
So this pathway corresponds to the top pathway illustrated in Figure 5, the numbers above the arrows being the genes involved. I’m curious as to what ways you could experiment to see if you could confirm steps in the process, or perhaps that’s not necessary so long as the end result is fluoranthene degradation.
The last sentence about P73T being potentially useful in marine oil spill bioremediation is a pretty cool possibility. I would imagine many more experiments and studies involving it would have to be carried out before it come be used practically, I imagine.
So while the researchers found these 138 genes that may be involved in aromatic metabolism, it’s likely not explicit that all of these genes are used in PAH degradation or fluoranthene metabolism. Could some of these genes hae been acquired via lateral gene transfer but otherwise not utilized (or utilized in other aromatic metabolic pathways not related to this study)?
Considering the age of this paper, have other significant groups of microorganisms capable of degrading aromatic hydrocarbons been discovered or have been the subject of greater focus in recent years? Are sphingomonads still the intense subject for researching PAH-degradation that they were when this paper was written?
So strain LB126 is especially significant because not only have the researchers identified the genes for fluorene degredation in a gram-negative bacteria, but in one that can sustain itself only on fluorine? And that makes it more significant than the pCAR3 plasmid from strain KA1?
So in order to test the expression of the dioxygenase-coding gene, they essentially constructed a plasmid by isolating the gene using PCR, subcloned it into the expression vector, and inserted that vector into the BL21(DE3) strain of E. coli?
I would guess that the purpose of dioxygenase overexpression is to grow colonies of E. coli with the vector containing the gene that codes for dioxygenase that would be able to express the ability oxidize PAHs, which would demonstrate that gene’s role in the process. Could you use gene knockout of that gene to reach a similar conclusion, like as was done in the previous paper?
Since they had to incubate the E. coli at 42 degrees C rather than the initial 37, are the proteins coded by FlnA1-FlnA2 in the E. coli different from those coded by the same genes in the LB126 strain of Sphingomonas because they have different activation temperatures.
I’m not entirely sure what the significance of the truncated orf2 encoding for a transposase. It apparently suggests horizontal gene transfer, but in the context of the whole experiment, it’s unclear to me why that’s important (or is it just an interesting observation?).
If both FlnA1-FlnA2 and CARDO can make 1-hydro-1,1a-dihydroxy-9-fluorenone from from 9-hydroxyfluorine, why is it that FlnA1-FlnA2 can make it from fluorine and not CARDO? Does this imply FlnA1-FlnA2 has a greater substrate range pertaining to fluorine degradation?
I suppose a closer reading of this paragraph actually answers my question on paragraph 3 about the importance of the truncated transposase gene. Is it the transposase’s proximity that leads them to the conclusion that LB126 acquired its genes for fluorine degradation through lateral gene transfer, as the paper only says it was ‘upstream’ (are they likely in the same cluster or group or something like that)?
Is it common for strains of Sphingomonads or other genera of microorganisms to use only a select few carbon and energy sources? Even if not, I imagine strains like A4 and the strain from the previous paper are novel since you only have one or two sets of metabolic processes to make observations about and detect enzymes and genes for.
Is the lack of reporting for the identification of dioxygenase genes for these sphingomonads due to just the lack of interest in identifying them, or is there some sort of significant difficulty in identifying them. If it were the latter, would reports exist that mention the challenges of identifying those genes (and the lack of said reports would indicate that difficulty is not the reason for the lack of identification)?
I guess this was their negative control to see if disrupting the arhA1 gene in A4 would diminish its ability to utilize acenapththene and acenaphthylene. By “blunting”, does that refer to removing the overhangs from the sticky ends of the DNA?
Is this preparation similar to what we did in lab where we used the oxidation of indole to determine which colonies did not have our inserts? I would guess it would serve the same purpose in this experiment.
Seems like the process of disrupting the arhA1 gene was effective. What is the putative arhA1 homologue mentioned referring to though? Is it referring to the gene they used in homologous recombination with arhA1?
While the arhA1 homologue seems to still be intact in the mutant, according to the results of introducing the mutant strain to the acenaphthene environment the strain couldn’t degrade it. So it’s interesting to consider what this homologue is. I’m curious as to what follow up research could be done to better characterize it.
So is the interest in the intrinsic electron-transport protein for ArhA1A2 sort of looking forward to the next step in PAH degradation, or is it another part of the initial process that hasn’t been well studied at the time?
In terms of bioremediation, does the Sphingomonad genus’s ability to degrade xenobiotics function similarly to its ability to degrade contaminants in the environment. Are ingested PAHs considered xenobiotics?
Primer extension is apparently useful in mapping the 5′ ends of RNA sequences. I’m curious to see what purpose this serves in the paper, since we never did primer extension (and I don’t recall if or how much we touched on it during lectures).
I imagine it’s common to use this many strains and plasmids pertaining to a particular species of bacteria in these studies. It feels like in comparison the number of strains me and my group have worked with pales (3 initial strains, plasmid vectors, and a few others). Of course, this paper probably took at least months of studies to complete, so if anything this would be what our research project would look more like were time not a limiting factor.
It’s a little sad that we never had time to do anything with Southern hybridization beyond discussing how it works, seeing how ubiquitous it seems to be as a method in these papers.
AG2-45, 2-48. and 3-15 lost the ability to produce indigo, but also maintained their ability to grow on 1,8-NA. Is that expected, or something not relevant to the conclusions suggested by these observations.
So it would seem the ArhA3 and ArhA4 genes were what the researchers were looking for in terms of the electron-transport system. It seems that incubation increased in cells expressing A1A2A3 without A4 (although not as great as with all four), so they’re complimentary, but are not both required for increasing acenaphthene oxygenase activity.
The arhR gene, interestingly, seems like a sort of “pre-step” for acenaphthene degradation, as its expression seems to affect the transcription of the arhA genes, and its disruption was shown to negatively affect acenaphthene degradation. It also seems to be active separate from the arhA genes.
So does this loose operon structure indicate an area for further research, or is it not relevant to PAH degradation since it doesn’t seem to have anything to do with acenaphthene degradation? Could it have been a result of mutation or gene transfer potentially?
I remember one of the papers (the first, I think?) involved theorizing a potential metabolic pathway in the breakdown of certain PAHs. They could potential do the same for future experiments. I agree that it seems difficult to accomplish with how dispersed they are, but I imagine there are ways we’re not aware of.
So since the arhA1 homologue’s function wasn’t identified, I imagine that finding its purpose would be a good jumping-off point for future research. Could you make preliminary assumptions of its function due to its homology to arhA1?
It seems so odd to me that despite the importance of Sphingomonas in PAH degradation we still know so little about their deoxygenation genes. It’s still somewhat unclear to me how this study increases our body of knowledge on the subject, but so long as it serves as a base point for further research, I suppose it doesn’t have to that much.
I remember during the oil spill, how the oil being released in to the deep ocean was causing big blooms in the bacterial populations that consumed the oil. this made big areas of hypoxias water as the oxygen was used up. and how this was one of the most damaging secondary events from the spill.
Its good how Antarctica is for the most part being protected from possible invasive species so we can keep it’s research opportunities available. It is interesting to see how these local environments are providing solutions for the problems we have created and how we have to be very inventive with finding these solutions since external fixes aren’t allowed.
I can see how our labs mimic this in our isolation of our naphthalene degraders. as our bacteria is isolated to single colonies to obtain strains for farther study.
PCR can be a highly powerful tool in the study of genomes as it can let us obtain as many copies of a piece of DNA as needed. most of the current Covid-19 tests are run through PCR to get the viral Genome large enough that it can be easily detected. which is also why it can take a while as PCR has to cycle through temperature ranges to successfully anneal and replicate the genomic material.
PCR and its readily availability has allowed for all the growth in genetic research over the last few decades as you can now easily get your hands on the equipment, proteins, and primers for replicating almost any DNA strain you want. I has need to all this testing with recombinant DNA and Plasmids as they are now cheap to produce once your initial sample is synthesized.
I probably wont be the three same strains but other bacteria in those areas can probably degrade oil. Oil and produces made from it are now everywhere thanks to humans, but oil will naturally enter different environments as it seeps up through the ground.
Life is a truly amazingly resilient thing. you can find some form of life at almost every point on earth. they have found microbes in salt water pockets at the bottom of mineshafts in Johannesburg. we have even done studies of exposing bacteria to space onboard the international space station, and found that several survived with minimal damage after an extended exposure.
Its interesting how they are using a naturally occurring environment with a significant amount of hydrocarbons as a base examination to compare to the areas effected by the oil spill.
Do these mats refer to biofilms or just large bacterial communities living off the oil on the ocean floor?
I see this group is also using 16S to identify and match what species of bacteria these are. but I don’t quite understand why they are using carbon 13 for this. is it just a way to mark the bacteria they could culture from the samples?
Its interesting that phenanthrene is the PAH that is mineralized the most. Especially with PHE being the PAH that was used in the last paper. Is there some reason, maybe with the origin of the genes that allow for the use of PAH, that make PHE a globally common PAH that can be broken down? maybe the proteins that allow for the breakdown were originally specialized to a different energy source that PHE manages to be the most similar to.
Its interesting that there are these specific genera that have fulfilled their niche so strongly that they are only able to get their energy from the breakdown of PAHs.
It will be interesting to see how they will test to find a coculture of bacteria. along with this how they verified if it is a symbiotic relationship, and how the bacteria handle these reactions
It is interesting that they couldn’t isolate the arsenite oxidase genes. is it possible that there is some temporary form the arsenite is being made into before it self oxidizes?
Since they all ready have primers for the genes dealing with the uses of arsenate and arsenite. I assume they are working off research that was already conducted before that had isolated the genes.
It is interesting the amount of sterility they are using to conduct these experiments. They are acting really carefully to prevent oxygen and sources of lab contaminates. The prevention of oxygen makes sense as the bacteria are supposed to be growing in a natural condition that lacks oxygen to use as the end electron receptor and are thus forced to use the arsenic.
Its interesting how few bacteria they found found in the samples and how only one close group photosynthetically oxidize arsenite. but this makes sense when you look at the graphs that the arsenite is being oxidized by phototrophic bacteria.
With the large rate differences between the oxidation and the reduction of As(III) and As(V). would it be safe to assume that in natural conditions, that the soil and surfaces in which the biofilm where collected from would primarily contain As(V) with very low amounts of As(III)
I wonder why there is such a focus on the salinities and the comparison to oil spills and leaks, is it that the two are linked or just that the risk is high in these regions of high salinity because of concentration of extraction sources or if equipment disrepair and misuse is the reason.
I wonder what they used as the initial cultivation media. Because the bacteria was found to degrade all of these hydrocarbons but you need a pretty specific initial media as to isolate the bacteria in the first place as too many added hydrocarbons and the media would just act as a general purpose media grabbing multiple different types of bacteria.
Okay, this answered my question from the first page, they used benzene as the carbon source in their selective media. As benzene is the primary structure for the different compounds in the BTEX group. It would make sense for what ever proteins the bacteria uses can also bind to the other compounds and break those down as an energy source as well.
Looks like they did the pretty standard practice of using Carbon 14 enriched versions of compounds you are trying to see the bacteria degrade. The amount of released Carbon 14 CO2 released can show the rate in which the bacteria is degrading the benzene.
Its interesting that the bacteria ceased to degrade benzene at 4 M NaCl, although 4 M NaCl is 233.8 g of NaCl in a liter of liquid. A this level of salt it is safe to assume that the bacteria began to shutdown its intake mechanism to help lower internal salt concentrations.
It is interesting that they didn’t manage to find all of the enzymes that would be used to breakdown these compounds. They believe that the enzymes should be known ones and not the possibly of unknown enzymes for the breakdown of these different compounds.
Its cool that they actually go through and show the actual breakdown steps for the benzene, toluene, and other hydrocarbons. It takes these different compounds and converts them to acetyl-CoA which it can then use in most metabolic functions.
This is looking a bit bit closer at the biochemical properties of the enzymes, they talk about the bacterial strain’s protein structure. The proteins are actively more soluble due to their abundance of negative charges. This helps the bacteria continue to function in high saline environments.
Fluorene is a know to be highly toxic to aquatic life. I found a document by the Minnesota department of health that talked about the exposure risk from fossil fuel contaminated areas and cigarettes’ smoke. They haven’t preformed wide scale testing but they believe it can build up in lake sediments.
This paper brings up that the fluorene oxidizing genes are on plasmids, this differs from the last paper where the genes were part of the bacterial chromosome.
Protein electrophoresis is harder than DNA electrophoresis as you need SDS in order to smooth over the charges to negative so they can be attracted to a single electrode so you can actually get coordinated movement of all the proteins in the same directions and at speeds equivalent to their size and not their charge.
Its interesting how they have the exact primers for these genes although this was one of the first times they were supposed to have been isolated for testing. There is no degenerate nucleotides in these sequences.
Its interesting how in figure 2, terrabacter sp. DBF63 had a regulatory gene in the reverse direction surrounded by Fluorene degrading genes in the forward dirrection..
Reverse transcriptase is such a powerful tool in our tool belt of techniques and enzymes for genetic research. We took an important part of one of humanities most infamous viruses and made it into a tool to try and improve peoples lives through the research avenues it has opened.
Its interesting that the Proteins produced, if they were FLnA1 and FLnA2, were not fully translated in their initial conditions, but when they were produced at 42C they got full sized proteins. I wonder what the physical/mechanistic reason for the translation to have stopped is?
I find it interesting that FlnA1 and FlnA2 could be a relatively novel way to break down fluorenone and other cyclic hydrocarbons, it would cool to see if the evolution of these genes could be determined to figure out how it came to be and function in this way.
Its interesting how this is a soil sample from deep sea sediment, this could mean that the ability to degrade these hydrocarbons comes from oil seep exposure.
That’s interesting how this bacterium is believed to have acquired its genes for the breakdown from 4 different sources.
cre-lox recombination is a very interesting way to inactivate genes. Although it is semi limited in its use as you need the recognized sequence for the enzyme to work.
I see that they used the now standard method of using mass spec and chromatography to detect the breakdown compounds made from the bacteria.
Its interesting that this paper actually when and tested all the different sugars the bacteria could use. It looks like they are doing a full examination of this bacterial genes/
Its cool how these little left over fragments of DNA can give an idea at how these different genes were obtained by this bacteria.
The inability of the knockout to use fluoranthene and naphthalene and the defectivity of the other hydrocarbons suggests that this was in fact the gene responsible.
I too have an interest in the differences between the inocula, or rather, what qualifies them as “very similar”. I can only infer that the differences and level of effectiveness will be further discussed in the remainder of the paper, for anyone reading would also pose the questions you did.
I too wondered why the bacteria were not isolated or investigated from this location. While this is the introduction and is meant to be brief, another sentence explaining the reasoning behind the absence of a study could have cleared up questions such as yours.
I understand the incubation temperature-wise of the culture, but am questioning the shaking of the samples. If it were surface level soil I would understand the need to recreate the chaos of an environment that is walked, run, jumped, etc. on but the article does not identify if the culture was taken from the surface or below. Why did they choose to do this, and why 100rpm?
What is the significance of taking the temperature at each site? I assume it is to collect data that is comparable between the different samples, but am unsure what exactly any temperature variation (or lack of) would signify.
Rod-shaped bacteria are significantly more common than other shapes in this study as well in historic studies. This paper provides evidence that it is likely due to the methods used in enrichment and subculturing. They also mention that rod bacteria can divide faster and have better contact with the surface. Does this imply that if the samples were taken from, let’s say, 2-3 inches down in the ground, that the majority of the bacteria would be cocci, spirillium, or other? If different methods were used in enrichment and subculturing, would the rod shaped bacteria still be most prevalent even if the sample were the same?
Figure 4 demonstrates that PAH concentration is reduced significantly after 15 days in incubation. Each chromatogram displays a spike between 15-30 minutes in regards to abundance before dropping back to almost 0 (x10^4). However, there is a slow exponential growth present at the end of each of these figures. Why would this be present?
Each isolate was identified by partial 16S rRMA sequences. Why did they use partial vs. whole sequences to identify? The similarities are mentioned shortly after, would the identifying partial sequences then be the portions that vary by isolate? If there was a 100% similarity between two isolates, should they not look at the whole sequence to make sure there is not genetic variation elsewhere?
Isolate LC is noted to be a human pathogen, and did not grow well in the study. However, the isolate had grown well enough to be sampled in one of the areas surveyed. Why then, was this sample not tested further? Many places where biodegradation is necessary are already hazardous to humans, so the addition of isolate LC, in an area where other isolates are not effective, could be plausible.
Hi Rachel,
I too noticed the reference to naphthalene. I do remember naphthalene was used in the experiments from the last paper we discussed, as a PAH tester. This coincides with the information you found via google.
With our previous research paper we discussed, it looks like hydrocarbon degradation in regards to PAH is under-researched in general. Because of this, I find your questions thought-provoking, especially the one regarding depth and temperature.
I also had difficulty understanding the “killed controls” at first. I believe that they were killing microorgansims, so that any changes can be attributed to the experiment at hand as Professor Ni Chadhain noted. I think they likely do so using one of the methods we recently learned about in our lectures.
Here it notes that only the replicate incubation with the fractions containing high amounts of DNA were used for analysis. I assume that this is to promote a higher chance of favorable data, however, would it change the nature of the data if fraction with lower amounts of DNA were utilized?
I was also intrigued by the method of neighbor-joining. From my understanding (as well as the professor’s comment) the largest advantage of it is the relative quickness as opposed to other methods. This would be interesting to compare to other methods we have used.
I believe that even though the presence of PAH degraders was confirmed, the prevalence of the degraders may not be large enough to show a significant improvement on the amount of oil. I think pursuing a way to amplify their prevalence/productivity could be beneficial.
I believe that the implication is that the research on this topic is still emerging, but I may be mistaken. Either way, the lack of genes characterized leaves area for exploration and a viable possibility for future research.
I googled the phrase “prosthetic constituent” and was unable to find a defintion. However, from context as well as the individual denotations of the words, I believe that it is referencing molybdenum, heme, and nonheme iron as components that together make up (along with the b-type cytochrome) the selenate reductase.
I believe that the antibiotics were added to all the media previously described, and to discourage the growth of bacteria that wasn’t intended.
I am also unsure about the exact purpose of analyzing the Se atoms, especially in so many ways. I do believe that this was performed after the degradation however.
I googled this and learned that cosmid clones are plasmids that contain the lambda “cos” sequence. It also appears they are commonly used in cloning, so their usage in this experiment makes sense.
This a question I was thinking myself. I believe that putting the cells directly into the environment would be the most effective way to start the reduction, but not the most cost-efficient. Introducing the organisms themselves would be both efficient and cost-effective.
Because the FNR gene regulates in such a way that requires lack of oxygen, the bacteria is best utilized in locations that present anaerobic conditions. This could be deep sea as in the last experiment, for example.
My question here is: how would one obtain further evidence that the FNR regulates selenate reductase during the transition from aerobic to anaerobic growth? Is this something that would be worth a future study?
I find it very interesting that certain bacteria or species can be individually identified using stain techniques and that those identified bacteria can then be grown on a plate in a lab. The most interesting part is what great new discoveries are in store due to techniques like these.
The first strain of Rhodococcus is capable of degrading several compounds that are harmful to humans and the second strain Rhodococcus sp. is capable of degrading some of these compounds at a higher level. I too wonder what the homological differences between the two strains are and I also wonder, how many other strains are there? What can they degrade? How many strains can be created using the original ones and what will they be capable of degrading?
Its interesting to see the temperature levels of the oven during the Gas Chromatography. I would have thought that a temperature of 300 ºC would kill the bacteria strain.
I wonder if they could also use TLC to see the process in which the PAH is degraded step by step using the hydrocarbon absorption?
I was wondering why they measured degradation at different concentrations. Is it in order to get a more accurate percentage and rate of degradation? Could it also be that higher concentrations of Fla allows the bacteria to mutate and become resistant to Rhodococcus sp. faster?
It is interesting to see that Rhodococcus sp. CMGCZ degraded Fla almost entirely, but ILCO is almost completely resistant. I wonder what structural differences between Fla and ILCO cause them to have such drastic resistance differences?
Im interested to know what future compounds can be created with rhodococci and which kinds of pharmaceuticals may already include it. I also researched that it is used in some herbicides which makes it interesting to see that something we have done as students in a lab can turn into something so useful for human life and development.
It would be cool to see the different substrates they could add to enhance degradation and know what specific effects they caused in the Rhodococcus as well as the bacteria it is meant to degrade.
We have seen that PAHs can be presence in both terrestrial and marine environments. Considering there are several ways PAHs can be transferred to humans, why do we not see more cases of PAH effects in humans?
I am curious to know the effects that the degradation of these compounds causes on the structures of the enzymes.
After reading the results section of the paper, it seems that the genes are similar but not exactly the same. This makes me wonder if the genetic changes occurred in order to degrade PAHs more adequately, or if the genes were altered as a result of some other evolutionary adaptation.
I find it fascinating that we use alcohol as hand sanitizer to kill bad bacteria and then we can also use a different form of alcohol to separate cells out to study them without killing them.
I wonder if the nucleotide sequence they found matched others in similar experiments. Also, it is very cool that they could have possibly had a different nucleotide sequence if they used chemicals from a different company or of different purities. Such small scale changes can have such massive effects in an organism.
After reading the results section, it looks like the sequence they found does match some other experiments findings. However, the sequences do not match up as similarly as the biologists here predicted. I wonder what happened that caused their prediction to be so different from what the experiment showed?
Tara, I had the same question. Do our degraders also have an aromatic oxygenase gene, or are there other genes that give provide the PAH degradation ability that our isolates contain?
It seems evident that these species containing genes similar to phnC have very similar evolutionary backgrounds. The differences in amino acid sequences make subtle enzymatic changes but the majority of the gene remains complimentary between species. It seems that the ancestor species expanded environments and its genes evolved making several different species depending on the available substrates in the environment.
I find it fascinating how this paper is so much different than the last paper even though they both are about the same thing. It is interesting to see the differences in how Cycloclasticus degrades PAHs versus Rhodococcus sp. and how two different bacterial species can use different mechanisms for similar activities.
Due to it having a similarity in amino acid sequence that is less than 44% and its cluster falling outside of the main cluster, does this mean that the phnA1 gene uses new proteins different from other bacteria strains to degrade PAHs?
I am interested to know why they were unable to get PCR products for the arsenic oxidase genes, what makes this gene unable to produce authentic PCR products?
It is cool to see that Arsenic is used by this bacteria similarly to that of PAH degradation by Rhodococcus and Cycloclasticus in that, both PAH degradation and use of Arsenic in the electron transport chain are mechanisms essential for survival of the bacteria.
I also wondered why they were stored in the dark. Could it be to prevent the UV rays from mutating the DNA? I would think this would not be the reason though because the bacteria live on rocks baking in the sun all day.
What is the purpose not specifying whether the tubes are shaken or not? Would it not be beneficial to either shake or not shake all of the tubes to ensure all tubes are prepared the same?
Paragraph 7 states that these results suggest that both As oxidation and reduction could be occurring simultaneously. How is this beneficial to the organism? Would it not be wasting energy by oxidizing and reducing the same molecules over and over?
The paragraph says that immediate oxidation/reduction occurred when the samples were in light or dark regimens for each transition. So, compared with the rate of reaction as well, what is causing some of the organisms to stop oxidizing or reducing the Arsenic? Are they finding new sources of energy that aren’t dependent on light/dark that they can use more consistently?
After reading through it again along with the comments, it is very cool that the different organisms are helping one another survive passively, simply by performing their needed metabolic activities.
The passage states that light-incubated biofilms abruptly ended activity at 50ºC. Is this due to processes like the heat breaking enzymatic bonds rendering the enzymes non-functional? I thought that biofilms were very stable and could endure more extreme temperatures than this so could it be some kind of defense the bacteria have that shuts down their activity?
I find it pretty cool that the bacteria can switch back and forth between photosynthesis and chemosynthesis in such short amounts of time.
Since anoxygenic photosynthesis is an ancient process, I wonder if the bacteria got their ability to do chemoautotrophy by random mutation influenced by natural selection, horizontal gene transfer, or another way.
[Samples were collected from non-diesel-exposed and diesel fuel-contaminated sites, and bacteria of interest were isolated by a three-step enrichment and screening approach, based on their ability to metabolize phenanthrene]
Later in the paragraph they mentioned that they were able to get three isolates that showed promising potential. How do non-diesel and diesel fuel exposed environments affect the bacterias ability to have a higher phenanthrene degradation ability? It also mentions that diesel fuel contains high concentrations of phenanthrene does that mean that there would be a greater chance of more degrading bacteria present in that sample from diesel contaminated sites?
[Diesel oil, the most commonly used fuel in Antarctica, contains a variety of toxic compounds that persist in contaminated soils, including heavy metals like Cd, Cr and Pb; and polycyclic aromatic hydrocarbons (PAH) like naphthalene, phenanthrene, and fluorene]
As I read this sentence, I wanted to know more on what exactly diesel oil is being used for. I found that people use it for everyday things such as making water, generating power for their heaters and lights, powering vehicles…etc. An enormous amount of energy (diesel combustion) is required to empower transportation due to Antarctica’s remoteness. It really puts into perspective how heavily they rely on the use of diesel making is seem not as easily replaceable with other options at this time. Would PAH- degrading bacteria be a temporary solution to this problem?
[Samples were sealed in sterile tubes and transported on ice to the laboratory and analyzed after 2 months.]
Is there a reason why they waited 2 months to analyze the soil samples? Would the time waited before analyzing have any positive or negative effects on the biological activity of the soil?
I found that in order for biofilm formation to occur there must be these three stages; attachment, growth, and dispersal. What could potentially be a reason for biofilm formation to not occur in this experiment? Does this have any correlation to the soil samples taken?
To add on, it could also have been due to the possibility of other PAHs, being used as additional energy sources, that were present in the mixture of the diesel contaminated soil that contributed to the result of not much of a lag phase and a high growth yield seen in S. xenophagum and R. erythropolis.
P. guineae had a high growth yield when isolated with phenanthrene as the sole carbon source and was found that this strain was able to generate robust biofilm growth in the experiment of phenanthrene stained crystals.
This was not the case when isolated with 0.2 % diesel fuel as the sole carbon source. It had one of the smallest yields. Since there is still phenanthrene present in the diesel fuel, could there be something present in the mixture of the diesel contaminated soil that might be inhibiting the bacterias growth in that environment?
I did some research on this and found that the increase in temperature increases the rate of oil degradation by bacteria. I do believe that this microbial bioremediation approach would be beneficial in other (warmer) environments where there are farms, industrial sites, landfills, and/or onsite sanitation systems.
I wanted to know more on in situ bioremediation and found that this technique limits the spread of contaminates from transporting/ pumping away to other treatment locations while the benefits of ex situ bioremediation include the ability to control variable temperatures, aeration, and nutrient level.
[Vibrio species commonly produce one or more siderophores to acquire iron ]
Siderophores are produced and utilized by bacteria as a result of iron deficiency. Are there other agents similar to siderophores that would help with other depleted nutrients in ocean surface waters such as phosphorus or silica?
[we identify other vibrio species that are natural aerobactin cheaters]
How efficient are these aerobactin cheaters? Since this allows them to conserve energy, would they, in most circumstances, out compete microbes that are able to produce siderophores?
I read that there is such thing as a solid CAS agar media that can also be used to detect siderophore production. How is the liquid assay more favorable to conduct in this experiment?
Could the use of mVenus to tag the iucABCD gene potentially alter its expression?
I believe it has to do with release of aerobactin in the V. fischeri ES114 culture fluids that causes competition within that co-culture. The siderophore competitively exclude V. harveyi growth since it does not possess aerobactin production genes. The siderophores act as a transport system.
I believe so. There was no competition between the two when there was enough iron present, V. fischeri did not have to release aerobactin and V harveyi was able to grow. This is not the case in minimal media. V. harveyi is not able to successfully compete with V. fischeri because it does not have siderophores, giving the advantage to V. fischeri ES114 in low iron environments. I also read that siderophores are able to complexes with other essential elements.
That also intrigues me. I would think that a species would only be able to acquire either or siderophore-producing and siderophore-cheating based on iron availability, not both. Wouldn’t this require a lot of energy? How could they keep up in these environments?
This is an interesting thought. I would assume that ultimately the species that were able to outcompete other species in low iron environments would at some point have to out compete what was once the most favorable such as being able to produce siderophores and siderophore cheating, leading to a new and more efficient way. Seems like an endless cycle.
I thought it was intriguing as well. Also interesting, how a wide variety of microbes must have evolved/ adapted to perform steps of the nitrogen cycle and use it as energy or nutrients.
I had a similar question. I read that even though OMZ’s can reach zero and become ODZ’s (oxygen deficient zones), “substantial suboxic nitrification has been reported in many of the world ocean’s major suboxic zones.”
Was this measured to give insight in what types of bacteria are able to grow in different areas of the water column based on temporal properties?
What could be affecting or contributing to the amount of manganese oxide present?
Figure 2 shows γAOB amoA expression being greater than Crenarchaeal expression of amoA
Why is the higher molecular weights of PAH’s used as growth substrates for soil bacteria and do the lower molecular weight PAH’s have the same effect?
In the introduction, it was stated that naphthalene is common in water pollution and has a poisonous feature that disrupts mitochondrial respiration. So the question I have regards the bioconversion of the pollutants in the water, did it cause any harm to those working in the clean up?
Why are the colonies phenotypically different and what bases were they chosen on?
Where was this strand of bacteria in relationship to the other 16S rRNA genes on the phylogenetic tree?
How is the emulsification activity and high cell surface hydrophobicity related to each other in this sense? Did napthalene emulsify because of its hydrophobic characteristics?
Looking at Paper 1- Fig 4, N7 had the strongest growth out of all of the strains compared to the others. I believe that this particular strain has qualities that naphthalene needs to thrive.
I had the same question, but I think that all of the proteins in the gram-negative membrane help with its tolerance and signal transduction.
Because only gram negative bacteria were isolated in this sample, is there a chance that if you go to the same area with a different type of pollution, would the results be different?
I agree with Gabriel on this, bioremediation, as opposed to using harmful chemicals, will help with the ecosystem immensely and could potentially aid in future oil spills.
The only question I have left is how much bioremediation will be enough to contain oil spills and still preserve the ecosystem?
How are these strains able to grow on petroleum?
Why are obligate marine bacteria only partially studied? What exactly does partially studied mean?
The SDS-PAGE will help break down the protein-protein disulfide bonds and will disrupt the tertiary structure of the protein. This brings the folded proteins down to their linear forms to help determine the sequence of the gene.
A biotransformation is a when a substance is altered, but what are they trying to transform the cells into?
If the genes for large and small oxygenase subunits are normally located in pairs, what does that mean for the PhnA1b strand that doesn’t have the small subunit?
Since the PhnA4 gene posses the similarity of chloroplast-type ferredoxins, the isolated bacteria will be able to act as an electron donor to nitrite, glutamate, and sulfite reductase.
The holoenzyme of an aromatic ring provides stability for the structure itself. Once the genes were introduced to E. Coli and expressed, does that mean that the phn gene can encode the holoenzyme by itself?
If PhnC is present, does that mean that the bacteria that was isolated is more efficient in breaking down PAHs?
Since PhnA has a wide substrate range, does that mean that there must be more regulation around that enzyme?
What is the significance of the reductase being coupled with dioxygenase gene clusters?
How is it possible that As(III) is able to achieve anoxygenic photosynthesis in photosynthesic bacteria? Photosynthetic bacteria are supposed to released oxygen.
What is the analogous coupled process that detects the redox reaction of As(V)/As(III)?
Why do you think that they were unable to obtain authentic PCR products for arseniTe oxidase genes?
Why did it matter that the slurries were stored in the dark at 5 degrees Celsius?
How does the annealing temperature effect the left of the fragment?
Why is a radioassay the preferred method of product detection?
Why do you think that there was a broader temperature range for anaerobic As(V) reduction than for the light As(III) oxidation?
Hydrogen is a better electron donor as oppose to sulfide. Is this due to fact that sulfide had large error association or because hydrogen is naturally a better electron donor?
Because the springs and ponds were shallow, do you think that if the bodies of water were deeper the biofilm would be less oxygen-dependent?
I’m a little confused here. What is the purpose of acetate in this experiment? In the previous paragraph it states that acetate does not stimulate As(V) reduction. In this paragraph it states that acetate is an electron donor. Does acetate just mark the presence of anoxygenic photosynthesis or does it do something more?
Since Rhodococci, can metabolize a wide range of compounds, does this mean they are found in more than one type of environment?
bioremediation can use microorganisms that are already present or introduced to the environment, does this improve the cycle of pollutants or does it somehow interfere sometimes?
what is cryic soil?
what is a white-rot fungus?
why did they wait until the 8th day? why not let the culture continue to grow?
how did the observers choose these time intervals?
why did the observers use two sets of degenerate primers after using bead beating technique?
would the results differ if they wouldve took readings at a different time in the experiment?
why did they use standard deviation in their graph, what does this explains?
what made them isolate the bacteria using phenanthrene
why haven’t the marine bacteria been observed as much as the terrestrial?
why was a taq dyedeoxy terminator cycle sequencing kit used?
what is sonication ?
is a partial ORF reliable as a souce as information as a complete one?
is looking at motifs exhibited by amino acids show homologous between them?
what does it mean when it formed a cluster?
what is the flanking region? and why is it so important ?
what is a diel cycle?
what is periphyton?
how did they decide what temperature to keep it at?
why was it prepared without nitrogen?
what was the point of the radiolabel experiment?
did they know that thioarsenic intermediates was soluble at high pH?
There are many different micro organisms that can be used. Some of these include Pseudomonas putida, Deinococcus radiodurans, Nitrifiers, and denitrifiers.
I had the same thoughts as Rhyann Davis. Is it proven that even though bioremediation solves the issue, it does not create new issues?
Is the Persian Gulf the only place this method is being used for pollution cleanup?
What is the purpose of transporting the seawater samples on ice?
What is the purpose exactly of the multiple changing temperatures?
This is probably a ignorant question but what are the red arrows on the table for?
N7 had the most growth at each increment but its peak alone was at 400 ppm.
I would think that all 54 strains were tested but as Sha’torrie said, only 18 strains showed enough growth to use.
Why would gram positive bacteria be more useful in this specific environment?
Narrowing the list down to the best degraders would save time for further examination and begin the process of finding which bacteria would be the absolute best for naphthalene degradation.
To me this sounds like the first experiments labeled certain naphthalene degraders. How did they do this without isolating these bacterias? Since the rest of is paragraph states that this second experiment is the first to isolate naphthalene degraders.
LB agar is known for shigella growth and plaque formation.
Subcloning is used to move a particular DNA sequence from a parent vector to a new vector. It could be used to isolate a certain sequence.
I would think the purpose for this is to get a more accurate reading of the ORF.
Horizontal gene transfer maybe?
Hydroxylation is the first step to oxidative degredation.
Why is strain A5 the only one without a plasmid?
Am I reading it correctly to say that the major cluster has a 44% level of amino acid sequence?
I would think that would be the case. It outed require more regulation to function as needed.
A diel cycle is a pattern of vertical migration common in copepods in lakes and oceans.
What were the first microorganisms found to be resistant to arsenic?
Wouldn’t this be the processes we discussed in class? Anoxygenic photosynthesis and chemolithotrophy?
Why would they need to run the experiment for as long as it was?
This is also why rivers have better oxygen levels.
I would think it would be because of the oxidation to As (V)?
That would make sense! It would definitely be easier for one membrane to adapt than two.
Actually, cooking seafood that contain PAHs increases the growth of the PAHs. Especially when the meat is smoked.
If obligate marine bacteria have only been partially studied, how do we know they will be better degraders than terrestrial bacteria?
Sulfide can act as another substrate for energy.
What is a alkalithermophile exactly?
I would think they would be less oxygen dependent because of the deeper water not containing as much oxygen.
On what grounds did they develop the hypothesis that this type of oxidation extended back that far?
Definitely more harm than good. As a species, we are terrible about exploiting and depleting natural resources with little to no ways of replenishing them. At the rate of the ice melting compounded with more pollutants, we may cause irreversible damage. At point what will we have gathered enough ice to prevent ecological damages? How will we ever conclude were to draw the line?
[Diesel-fuel contains high concentrations of two- and three-ringed PAHs, including phenanthrene. Since it exhibits higher solubility than others PAHs in water ]
It would appear that perhaps the combination of the high concentrations and higher solubility of the diesel-fuel contaminants assists the bacteria as there would be an abundance of resources for them to feed from. As for your second question, I am not so sure myself. Over the summer, I got into brewing alcoholic beverages. In my research, I found out that too much sugar could have negative affects on the yeasts. Granted, yeasts are from another kingdom, but the same biomechanics would apply to bacteria.
On my last sentence, I meant to say, “same biomechanics might apply to bacteria.”
Luria-Bertani broth supports a quick and steady growth, however, it is carbon limited. This means the population will plateau quickly, and then the population will decline.
With room temperature being optimal for growth, I would imagine that temperatures warmer than room temp, would cause the bacteria to a bit too quickly, possibly hindering the chemotaxis assay. Some growing colonies may block pathways for other bacteria trying to swim to the chemoattractant substances.
Figure 5 shows how the deletion of iutA or fhuCDB genes causes the two strains’ growth to be completely hindered. What is it about these genes that makes them essential requirements?
From what I gathered in figure 5, it appears that all of the genes are necessary. It’s highly likely that they all work together and missing just one could completely inhibit the strains.
They are more important for the reason of being able to fix nitrogen in an extremely low oxygen environment and they account for more than the “usually less abundant AOB.”
The anoxic layer has more to do with differences in water density. Since both layers never mix, the anoxic layer maintains a low oxygen concentration while the upper layer is free to cycle oxygen.
I’m not sure if it’s the default method, but I would guess so as it’s a highly accurate method to detect, identify, and count specific cells.
Like many instruments, flow cytometry count cells by measuring both visible and fluorescent light.
In fig 2 b and c, there seems to be a correlation between the concentration of O2 and transmission spiking nearly at the 20 meters mark.
I believe so. High concentrations of naphthalene are toxic. The data seems to reflect that too little and too much naphthalene affects the growth. I would think they could isolate the ones that thrived above 400ppm and harness their degrading abilities. That way they can have a understanding of the bacteria that can handle pretty much any situation dealing with the degrading of naphthalene.
I am glad that organisms exist that can help degrade oil pollution in the waters. Until this class I did not know that there was such a thing. Is there current research on weather or not other organisms can be introduced to the affected environment to help further degrade the accumulated microbial biomass once the naphthalene is degraded?
It would be interesting if we could introduce the degrading bacteria into the diet of the aquatic life in the affected environments so that there will not only be a constant supply of the degrading bacteria but also so that the other organisms could potentially keep reproducing these bacteria. Is this possible?
I believe the samples need to be shaken throughout the incubation in order to ensure there is no settling of the particles. If they are constantly agitated then there is very low chance that a portion of the sample was not given the same amount of exposure to the NP degrading bacteria. Therefore reducing the chance of the data being skewed.
In order for PCR to work high temperature must be used to eliminate hydrogen bonding between bases and the newly synthesized bases. You need to denature the strand(high temp), anneal the primers (lower temp), and then allow for synthesis to occur and then repeat the cycle.
Since this is the first report on isolation, have these selected bacteria always possessed this ability to degrade naphthalene, or is it something that they were able to quickly develop since it became a rich carbon source that was not always present in these concentrations?
It looks as though BH medium is used to isolate microbial hydrocarbon deterioration. only those able to decompose hydrocarbons will grow. It is designed to add your hydrocarbon of choice for your particular study, which is perfect for this experiment.
Im trying to recall from organic chemistry but is some form of halogenation the way that the phenanthrene can be removed by trichloromethane?
Was the antibiotic that was selected something that the E. coli, were resistant to? Therefore making it a substance that the microbe would like to take up and use it for its own benefit?
Chemolithoautotrophs are organisms that obtain their energy from the oxidation of inorganic molecules and it typically aerobic. Photolithoautotrophs use light and an inorganic electron donor. These would be the organisms that could use the toxic form or arsenic for their energy source, correct? I assume are the two types of organisms this paper will be focusing on based on the next four paragraphs.
Are they not focusing on oxic conditions because is has already been manipulated in lab? I think they may be doing so because metabolism under those conditions may be too predictable and not yield a very “interesting” result.
Since their focus is on the anoxic conditions, I think they are trying to see is these organisms are able to give insight on how things were way before there was atmospheric oxygen. I believe arsenic is so toxic to use because we are dependent on oxygen and so maybe this study is trying to reveal how organisms operated before oxygen was even a possible to be included in a metabolic process.
Based on the figures about 45 degrees Celsius is the optimal temperature for maximum As (III) oxidation and As (IV) reduction. This would account for the abundance of the red springs, correct?
Did they decide to disregard the green samples? I do think that the red samples will give out more results to work with since they exist primarily at the optimal temperature for the strain. And I think they have plenty of experimental information from other studies on the cyanobacteria.
I was thinking the same thing. If the DNA responsible for the degradation is in a plasmid that we can isolate then we should be able to genetically modify similar organisms that may not necessarily be able to degrade PAHs. We could then recruit many other organisms and use them to our advantage in this case.
Once the DNA sequence for the degradation of the phenanthrene is obtained could it then be introduced to organisms that can only breakdown NP? So if these organisms were introduced to an environment that contained both, could it then degrade one after it is done degrading the other?
Are there ways to amplify the production of catechols in the cell since the are essential to breaking down PAHs? If so could you introduce different gene sequencing or other non harmful chemicals to the environment to increase this production?
Do these specific pathway make the strain have more affinity to degrade phenanthrene vs the PAHs? Or does it degrade any PAHs in its precense?
Would introducing the genes responsible for the meta-pathway into strains that can thoroughly degrade NP in order to create the perfect PAH degrader?
I would like to see if we could mimic these chemical pathways and enzymes out side of the cell so that we can introduce the batch of chemicals to a contaminated area without have to rely on a living organism to help bio remediate contaminated environments.
Could the introduction of indole to the contaminated environments encourage the strain to degrade more rapidly, almost like a bait?
In the figure the oxidation of As(III) and As(V) reduction are proportionate to each other through out the time of the experiment. Arsenite being the electron donor, and arsenate being the electron acceptor. Arsenate can be used as the terminal electron acceptor. Starting off in the light Arsenite oxidation is favored and in the dark arsenate reduction is favored.
It looks like they wanted to show that using a different carbon source this one being acetate, shows that the organisms do not favor oxidation in the dark at all.
Since these oxidation and reduction reactions are occurring in the same place then there must be a mixture of organisms that are metabolizing these species of arsenic. In the light it is likely that it is an aerobic phototroph and in the dark it is most likely a chemolithotroph.
Could it be that the redox potential for H2 and H2S is higher than that of Ar, that causes the organisms to prefer to use the Ar species as their electron acceptors and donors?
I believe that since the microbes that are reducing in the dark are non photosynthetic then they are not relying on light to carry this out. In the light the temperature is usually in a smaller range than in the dark, this is my best guess.
That is very interesting! I did not know that saline liquids (think fracking) are used to extract hydrocarbons. This is a very important and interesting research topic as many well-studied PAH and BTEX degrading strains do not tolerate high-salinity environments. The high salinity could prevent many strains from growing and ultimately bioremediate and so identifying halophilic and halotolerant organisms is crucial for bioremediation in salty environments. Mannitol salt agar can be used to select for halophilic organisms.
That is an interesting point, Avril. I wonder if we helped efficient bioremediating halophilic strains by adding nutrients etc., if also beneficial compounds would be degraded. For them both are just food sources. Also, some strains may be (opportunistic) pathogens that could potentially harm the environment and the animals living in it. I guess we would have to look at the pros and cons and evaluate.
That is a good question. Maybe start with an enrichment culture which could enhance the density of organisms that could utilize phenol as an energy source for example and then use mannitol salt media to determine if it was halophilic or halotolerant. In this paper, the sample was collected from a salty and oil impacted area so it can be assumed that they can degrade hydrocarbons and are halophilic but it would need to be verified. When it comes to the individual compounds that the Arhodomonas sp. Strain can degrade I think they would have had to test this after the isolate was isolated.
Luria Broth is a nutrient rich medium often used to grow up a variety of bacteria. Minimum salt medium does not contain a lot of nutrients, but we added indole and naphthalene making it a differential and selective media. The conversion of indole to indigo (blue colonies) indicates the presence of monooxygenase and works as an indicator of PAH degradation abilities. Due to the lack of nutrients, only the individuals capable of utilizing naphthalene as an energy source were able to grow up. I am not 100% sure but they may have used both MSM (supplemented with 1 M NaCl) and Luria Broth to see how well the isolates grew in an environment with a higher salt concentration. They knew that they would grow well on Luria Broth but MSM with NaCl would give additional information. Finally, they tested the benzene degrading abilities of individual isolates from both mediums.
I found that they used a GC equipped with a flame ionization detector and a DB-1 capillary column. They also used helium as both the carrier and makeup gas. I think that it is pretty cool that they set up microcosms and used the production of 14CO2 to study the strains’ ability to degrade benzene under various concentrations of NaCl.
I would have liked to know how many bottles containing benzene and NaCl there was and at what concentrations. I only know that it ranges from 1 -2.5 M Na Cl. Also, is 24 micromolar the minimum and 34 micromolar the maximum degraded within 2-3 weeks in any given bottle? We know that the benzene degradation ability was tested from 0-4 M NaCl with no growth at 4, 0 and 0.5 NaCl. The maximum degradation rate was 2 M NaCl which is good to know when evaluating bioremediation strategies in very salty environments.
It is interesting that the Seminole strain can degrade benzene and toluene but not ethylbenzene and xylene as these structures are very similar. More details about the degradation pathway are needed to figure this out. The isolate might be lacking one or more necessary enzymes to catalyze certain conversions.
I like that they measured the growth of the Seminole strain in addition to quantifying the benzene degradation. That way they can be pretty confident that they are growing by using benzene as the energy source.
I wish they would write a sentence about how efficient strain Seminole was at degrading phenol. I think it is pretty cool that the degradation results match the genome analysis. Could not grow on Benzoate, GA, and hexadecane since no specific enzymes catalyzing the initial step of these compounds was found. But what about CAT? Were such enzymes found but still no degradation was observed?
I see now that part of the information I was seeking could be found in the tables.
The one sample t test looks at two means and determines if they are statistically different from each other. In this case, it was used as a tool for comparative genomics between the Seminole strain and 10 other hydrocarbon degrading halophytes. They found 3 COG categories that were significantly different. This method seems very straight forward and I hope to use it in my research project.
Upper and lower degradation pathways denote 2 different routes that degradation of a compound can take depending on which enzymes that are present. For example, in the upper deg. route of benzene the enzyme epoxide hydrolase adds a water to the epoxide making a dihydrodiol. Further down the pathway the muconaldehyde is converted to muconic acic which is actually a common biomarker for benzene exposure.
It is incredible that we are able to use bioinformatics to predict gene functions based on codon similarities to previously described genes. In this case, after proteomic analysis, which looked at upregulated genes while exposed to toluene, 12 gene products out of the 19 ORF’s predicted to be involved in metabolism of toluene was indeed significantly abundant. It is not ideal that some of these genes were slightly upregulated in the negative control, but they had reasonable explanations for it.
I believe bioremediation definitely could contribute to degrading and removing some of the pollutants you are talking about. Although, it is important to keep in mind that there are hundreds of PAHs and there is not one strain (that I know of) that can degrade all of them. Some bacteria will degrade some PAHs better than others and a mixture of isolates are usually most efficient. That would be an interesting location to do toxicity testing and other experiments. We could for example improve the bioremediation by adding nutrients.
What does an angular dioxygenase attach? From figure 1 I can see that 2 hydroxyl groups are added to carbon 1 and the carbon left of it (1-hydro-1,1a-dihydroxy-9-fluorenone). It is interesting that Arthrobactersp. strain F101 posses the genes to degrade fluorene necessary for all the 3 pathways discussed in this paper. Sphingomonassp. strain LB126 only utilizes pathway number 3 (to the right in figure 1).
It looks like this article describes many useful experiments that are very applicable to our class. However, a lot of the procedures have been conducted and described before and so this paper cites them. For example, plasmid DNA extractions, restriction enzyme digestions, ligations, transformations, sequencing, and agarose gel electrophoresis were carried out using methods described by Sambrook, J., E. F. Fritsch, and T. Maniatis. 1990. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. Thus, to get the methods we would need to follow the citations and find the primary research and publications.
I did not know what primer walking was, so I googled it. It is used to clone a gene from its known closest markers. After sequencing the first piece the following piece is sequenced using a complimentary primer to the end of the first piece. Also, having a primer table instead of writing out the sequences in the previous paragraph may have looked cleaner and better.
It is surprising that no primers could amplify conserved domains of previously described PAH dioxygenases but great that dibenzofuran 4,4a-dioxygenase was identified from a gram-positive bacterium.
I did not know that a transposase suggests that the adjacent gene cluster was acquired by horizontal gene cluster, but I guess that could make sense. orf2 encoded a truncated transposase but no change in GC content was noticed. Definition for a transposase is that these are enzymes that identify the inverse terminal repeat sequences within the DNA and proceed to bind and excise the DNA transposons in between the terminals.
This paragraph was difficult to understand. Could you explain it a bit during class?
I had to read this paragraph many times to make any sense of it. Table 2 essentially shows the substrate range of FlnA1-FlnA2, from Sphingomonas sp. strain LB126 as expressed in E. coli, and it was compared to 2 well-studied angular dioxygenases abbreviated DFDO and CARDO. When fluorene was the substrate these are the 3 oxidation products: Dihydroxyfluorene, 1-Hydro-1,1a dihydroxy-9- fluorenoned and Fluorenol-dihydrodiol. 1-hydro-1,1a-dihydroxy-9- fluorenone (63%) was the main product and was identical to that of the DFDO mediated oxidation product of fluorene and the CARDO-mediated oxidation product of 9-hydroxyfluorene. However, CARDO does not yield the main angular dioxygenation product when fluorene is used as a substrate which was expected. All the products produced when the substrate was naphthalene and biphenyl also formed by DFDO and CARDO.
After reading this paragraph again after today’s lecture I understand it much better. I am sure it was tough to see flnA1 give a slight band when grown on glucose. I am sure the next phase took quite long as the authors had to repeat multiple steps to find the ideal growth conditions yielding a larger proportion of the FlnA1 and FlnA2 proteins in the soluble fraction. Finally, to confirm the results no oxidation products were detected in the GC-MS when a strain without these genes were grown under the same conditions. This method seems time and labor intensive but very fascinating.
Growing Sphingomonas sp. strain LB126 on other similar compounds is a great way to assess substrate range of this pathway. So, when grown on dibenzofuran it was transformed into 2,2,3- trihydroxybiphenyl by FlnA1-FlnA2. Is this the first step of was it first transformed into the unstable hemiacetal? Angular oxidation products were not detected on either carbazole and dibenzothiophene. The activity of A1-FlnA2 may have been too low. Another referenced paper also found that their isolate could not perform angular dioxygenation of carbazole either.
It is always interesting to research something has not been studied much before like PAH degradation by marine bacteria. I wonder if there is a specific reason for isolating a bacterium from deep sea sediment from the Indian ocean. Had the area been affected by an oil spill maybe. We are probably going to recognize some of these methods from the first two papers.
I could not remember what non-alternant meant so I googled it and it is π-Conjugated carbocycles containing odd-membered rings. High molecular weight PAH is generally more challenging to degrade so I am interested to see how efficient C. indicus can degrade it.
Illumina Solexa sequencing yields short reads up to 100 bp. PATRIC predicts protein coding sequences after annotating the genome. Island Viewer only accepts genomes in GENBANK and EMBL format. Following annotation, we receive a document in embl form. Could we use this on the island viewer website?
I think gene knockout would be better to use if you do not know the function of some novel gene. It is a great confirmation experiment by looking at the difference with and without a gene. However, I feel like RT-PCR is easier and less complicated to do. I would feel more confident that I have found the gene of a certain function if I did a successful knockout experiment. Partially because previous papers have mentioned various reasons for why a gene appears upregulated when not expected to. Leftover mRNA etc.
G+C content can be used to identify regions of horizontal gene transfer. The G+C content can vary from 25 to 75%. The complete genome of C. indicus P73T is 65 mol% and contained 4827 predicted protein-coding sequences (CDSs). It is odd that no gene for tRNA-Tyr was found when the corresponding tyrosyl-tRNA synthetase gene (P73_1712) was identified.
Figure 1 displays the Circular maps of the chromosome and five plasmids of Celeribacter indicus P73T. I like the circle for the chromosome but the circles representing the plasmids are so small that they are difficult to interpret.
It is very interesting that 37 GIs were identified by IslandViewer and it definitely makes me want use it for our project. Can PATRIC identify horizontally transferred genes (HTG) in our bacteria’s genomes? It is interesting that non-essential genes such as PAH degradation genes were acquired through HTG and is likely helping strain P73T’s ability to degrade PAHs.
The plasmids do not contain a lot of DNA. All five plasmids together make up about 9% of the total CDSs in the genome. The arrangement of four COG functional groups were quite different in each plasmid indicating multiple origins and a specialized function for each (PAH degradation).
rep and parA genes were used to do a phylogenetic analysis. I did not know what parA was, so I googled it. It is responsible for plasmid partition. It ensures the proper distribution of newly replicated plasmids to daughter cells during cell division.
I am surprised by how many ring hydroxylating dioxygenases the genome contains. These proteins are multicomponent bacterial enzymes that catalyze the first step in the oxidative degradation of PAHs. Is having multiple genes code for the same enzyme going to make the bacteria better at initiating PAH degradation?
It is clear that strain P73T have acquired genes through lateral gene transfer as region B of the genome is absent in another bacterium of Celeribacter strain B30.
Despite the number of PAH degrading genes found, only the C-7,8 dioxygenation pathway involving extradiol cleavage of 7,8-dihydroxyfluoranthene was used. Thus, strain P73T can be useful to study this pathway in particular. I think extradiol cleavage enzymes incorporates one O2 next to one of the hydroxyl groups (not in between) and activates molecular oxygen.
Since bioremediation is the best restorative means to combat PAHs in ecosystems, does it permanently restore those ecosystems back to their full potential or does it temporarily revive them? Also in regards to Fluoranthene being carcinogenic, what disadvantages are there to using it?
What are other supplemental inorganic nutrients that enhance PAH-degrading microorganisms? Also does having low amounts of YE and glucose limit the degradation potential?
Tim,
I was wondering the same thing initially. However, now that we are writing our summary I am under the assumption that these nutrients don’t always enhance PAH degradation compared to it’s normal ability.
What is the purpose of re-suspending the Fla grown culture into MSM prior to inoculating at OD600? Does this enhance the growth?
What is the main difference in using two different enriched culture ratios in YMSM? Why was the 1:50 ratio chosen to maintain the enriched culture every 16 days instead of the 1:10?
Justin,
I think that the purpose of spraying a Fla solution on the agar plate is to test whether the strain CMGCZ will degrade Fla at all.
Why was the degradation such a huge jump between the Fla and the Nap/Phe PAHs? I would have assumed there to be a difference in degradation between Fla and other PAHs but not this significant.
It’s interesting to me that there is no degradation delay in the YMSM medium while the MSM medium delayed degradation by 24 hours. I wonder if the pH, cellular transport properties, and/or chemical structure play a role in the rate of degradation?
It is evident that clear zones forming on MSM agar plates is a technique used for identifying hydrocarbon degraders. But is this technique the best suited for screening or are there other methods that show better identification?
It’s unique that rhodococci can be applied in environmental remediation, in pharmaceutical, and chemical plants. But what is the overall sufficiency of any PAH degrader and is it worth utilizing?
Since Cycloclasticus contains a variety of aromatic hydrocarbons do we most commonly or only introduce this bacteria to marine environments today? Is this the most productive biodegrading bacterium?
I wonder how often there are found cases where a person has experienced life-threatening symptoms by consumption of seafood contaminated by PAHs? Is there a method for seafood-distributors to screen for PAH contaminants prior to market?
Aubrianna,
Honestly this has been resonating with me from the beginning as well. You hear precautions on just about everything else through the news and commercials. Though I have never heard anything regarding PAHs.
Even after completion of reading both papers 1 and 2, I am still not certain which strain of bacteria would have a more beneficial degradation ability.
What is the purpose of noting where the chemicals were manufactured? Is it necessary for other researchers to use these exact companies to replicate this experiments results?
I was thinking the same thing when I first read about their use of the lac promoter. I also was trying to visualize how they would use lac operon’s activating and repressing abilities to help them with their study.
I find it interesting that the amino acid sequence of PHnA1 showed a 51 to 62% identity match with the sequences of PAH dioxygenases while other subunits showed a much a lower identity match. What would be the major factor of this conclusion?
Will PAH degradation decrease since there was no small-subunit gene found in the flanking region of phnA1b and typically oxygenase subunits are in pairs?
Jesse, that’s a great observation! I never thought to think that varied gene location in these species could relate to different habitats. This could strongly correlate that one may have a greater PAH degrading ability over the other.
Lameace,
I too agree that this seems like a much different result than our first paper. PhnC seems like it would be a much greater degrader of PAHs both LMW and HMW.
Lameace,
I too think that this is interesting considering we know the extreme toxicity that arsenic, arsenate, and aresenite bring to the environments. I am interested to know how can bacteria utilize those oxyanions that are so toxic to create more energy without dying off?
Since the researchers were not able to retrieve viable PCR products for aresenite oxidase genes could that mean that the wrong PCR conditions were used or possibly the DNA template has degraded?
Jessica,
I was wondering as well why exactly did the researchers choose to pursue only the cycling of arsenic under anoxic conditions?
Derria,
Periphyton’s are simply underwater organisms that attach to surfaces of some matter.
Hang,
I agree that the environments characteristics can lead to vital information for researchers to discover certain metabolic reactions. I also researched more on other reasons that scientists believe the habitats are of importance. A new idea is that specific environments are playing a major role in antibiotic resistance globally.
Jesse, I was wondering if the light vs. dark incubations have anything to do with motility abilities.
I think it is strange that the temperature ranges were very different. How could the dark As(V) reduction occur over such a wide temperature range while light As(III) oxidation happened over such a limited range? I would think that they would have more resemblance in their temperature ranges.
How could researchers further test to confirm or disprove that the result correlated with the inefficacy of the primers or due to an aoxB-independent mechanism?
Jessica, I too was wondering why the temperature range wouldn’t be consistent between the two. Also, I am curious to know what other environmental changes and/or metabolic changes could be responsible for driving light and dark oxidations/reductions?
Jennifer, I was wondering the same thing. Is it an environmental factor or a metabolic function that is causing acetate to have no known effect on As(V)?
When measuring the biodegradation ability of the naphthalene degrading bacteria, is the speed of the process or the purity of the end product more sought after?
I would hope more areas used this process due its more environmentally friendly nature.
It’s always great to see research that is trying to make the world a more ecologically self place. Future research in these areas will help with any future oil spills.
The fact that only Gram negative bacteria were found is interesting. What would the environment look like for it to have only been Gram positive bacteria? Is that even possible?
A dendrogram is “a tree diagram used to illustrate the arrangement of the clusters produced by hierarchial clustering”, according to Wikipedia. They are commonly used to illustrate the clustering of genes.
It’s interesting to note that the researchers did not find a significant function for orf7 and no obvious homolog and the fact that it is the only ORF being read from right to left. The rest of the found ORFs are read from left to right.
What other redox systems specifically would the electron transport proteins of the PAH dioxygenase systems share with?
What is the significance of examining further the combining of PHnA1b and PhnA2?
What determines whether the uncontaminated soils from going through As(V) reduction or As(III) oxidation upon arsenic oxyanion amendment? How it uses arsenate as an electron accptor or arsenite as an electron donor?
How does using As(V) as a respiratory electron acceptor be used to employ inorganic substances such as sulfide or H2?
Is there any significance for shaking or not shaking the tubes being prepared for aerobic incubation experiments?
What is the criteria for terminating the experiment? Experiments were stopped after 2 to 7 days.
A halophile, according to els.net, is a “salt-loving organism that flourishes in saline environments”. The Archaea found in the 16S rRNA gene clone libraries are all related to extreme halophiles and are likely present due to the high salt content of the Paoha hot spring environment, as the paper states.
It would be interesting to see any adaptations in electron donors from the biofilms if the hot springs were to ever change or become polluted to the point of mutations being necessary to survive.
So this is one reason why there is no life on Antarctica because of these pollutants. Earth is in danger because these pollutants could are increasing global warming.
Is there a way humans could travel there without causing such pollution? Are small invertebrate organisms decreasing now?
Using microorganisms sound like an effective way to help keep the soil of Antarctica in good condition. Wonder if there were different approaches before the idea of using bioremediation.
The ex situ process seems like a better, but longer and time consuming process. The in situ process seems like an ineffective process. Are there any major differences between the two when deployed and which is being utilized most often?
What other group of bacterial strains from these communities have been found to aid in bioremediation? How many hydrocarbon degrading bacteria have been found so far?
What was the purpose of taking samples from non diesel-exposed control sites? Perhaps to gather a statistic of the different phenanthrene degrading bacteria in those areas to the diesel-exposed sites.
So biofilm was used as an indicator to stain the cells of the phenanthrene degrading bacteria? A scanning electron microscope had to been utilized to view the cells’ surfaces. Was this process a necessity since a phenanthrene adhesion assay was done?
What does an unexpected higher level of fluorescence indicate? Is there a specific emission peak range when detecting bacteria contamination?
So the phenanthrene degrading bacteria can use diesel fuel as a carbon source but doesn’t have to show any chemotactic response. Why?
I wonder what the effects of bioremediation would have been like if foreign organisms were allowed and if the process would had been harder and longer to accomplish.
I wonder if scientists could ever balance the recurrence of human activity whose to blame for these PAHs and keeping the numbers of these pollutants low.
Would the cultivation dependent approach be faster than the cultivation independent approach?
I found out that petroleum is the largest oil source in oceans. Does the Guaymas Basin contain the most petroleum hydrocarbons of all oceans?
So caesium chloride gradients help the DNA remain undamaged and allow DNA densities of 3.99 g/cm^3 to migrate towards the top. Ethidium bromide is the dye that makes DNA appear as fluorescent bands.
I wonder if mineralization would have occurred in the 4571-2 sediment if the temperature would have been increased.
I wonder why labeled 14C-labeled PHE did not start growing until 3 days after the unlabeled substrates.
Why is heavy DNA and light DNA utilized in SIP-DNA? I understand the different densities.
I wonder if there were other methods than SIP-DNA that could have helped with the identification of microorganisms in deep-sea sediments.
Not sure if it was stated in the article but why were unlabeled PAHs used?
I wonder if As(III) oxidation and As(V) reduction would function better and quicker in anoxic or oxic conditions.
I wonder if the anaerobic As(III) oxidation and aerobic As(III) oxidation would be better metabolites than As(V) reduction in oxic conditions
I wonder if the same construction and analysis was conducted for the green-colored anoxic spring.
I wonder why, at a higher temperature, more base pairs were produced with a smaller band size.
I wonder if oxidation and reduction rates lowered after 200 h of incubation because of low H2 concentrations.
Maybe there were too many differences in the sequences that resulted in PCR error.
Anoxygenic photoautotrophs like cyanobacteria, helped influence this biogeochemical cycling.
I also wonder what novel mechanism would be used to detect arsenite’s oxidase genes.
What’s the purpose of using labeled PHE and unlabeled PHE for SIP incubation?
My question is if these bacteria are able to help reduce the pollutants produced by diesel fuels would we be able to use the bacteria in other places in the world where the population is denser and an extremely large population is? Also would these bacteria be able to survive in other ecosystems or would they have to be adapted for specific regions of the world?
I agree that for years and years mankind has been causing harm to the planet but I believe this was due to the ignorance that we had about the science of the world. Although humans are the most advanced species on the planet we don’t have the answers to everything but as we find things out I think we do try to make changes in our shortcomings. For instance, in our advances in electric vehicles rather than continuing gas powered cars.
What does it mean for the bacteria to be grown with agitation? Is it what it sounds like in the fact that they just shake/stir the samples?
How would they ensure that the bacteria were not exposed to any diesel in the control sites. I assume that since the antarctic is extremely cold it probably would not be a very viable option to travel without some sort of transportation and also if they did manage to travel without a vehicle since Antartica is fairly desolate wouldn’t all of it have the same exposure to the diesel?
I imagine it just like with any other living things. Certain requirements have to be met for that thing in order for it to grow. For instance some plants can only grow at high altitudes and some at low altitudes everything has a sweet spot.
I find it very satisfying that this strain of bacteria shows promise in the fact that it can potentially help clean up the mess we’ve made with the use of fuels like diesel. With the help of these microscopic organisms we can make our home clean again.
I understand that these bacteria are specialized for cold weather regions since they were found in Antartica. But would you be able to manipulate these bacteria to continue working in all climates? I think the possibilities for these bacteria could be much larger than just cold weather climates.
Yeah for such a desolate place it is amazing to know that there are still so many different kinds of life even though they are so small. I wonder if finding life in such a cold place could alter what qualifications a planet should have in order to find life.
How do they study these samples of bacteria? Meaning since they come from the deep sea and are adapted to these high pressures as well as temperature how do they transport and gather these samples?
The exploration and understanding of new microbial oil degraders could help us find one that is more efficient at degrading oil and could help with clean up of major spills as to not cause a major impact on any ecosystems.
I was wonder do micro organisms do well with extreme changes in pressure? For instance in this study it says they took the samples and immediately sent up.
I see that in this paragraph they seem to be doing what we were doing in lab a lot with basically trying to get samples from cultures that had positive results. They took these samples and stored them much like we did but why did they end up freezing them at -80 C was this also part of the test or was this just a way to store them for use later?
“The vent fluids, which are laden with petrochemicals, migrate upward to the sediment surface, thus providing a natural model system for studying the microbiology of deep-sea hydrocarbon-degrading communities.”
This is a portion they stated in the introduction describing why they chose this area. Not necessarily due to frequent ships but because of the naturally occurring communities growing there.
I am sure there are other bacteria out there that would hold similar capabilities as Cycloclasticus. Especially due to the fact that there is still so much of the ocean that has not been explored. On top of that micro organisms are not visible to the naked eye so knowing where to look for these things just add to the complications of locating similar organisms.
Vila, J., Tauler, M., & Grifoll, M. (2015, February 03). Bacterial pah degradation in marine and terrestrial habitats. Retrieved March 12, 2021, from https://www.sciencedirect.com/science/article/pii/S0958166915000154?casa_token=AnAEB0nP4x0AAAAA%3AdMDKCrUEKli8JOEgEyw450L3_sZLfkTwMKxLgC25nPndYcNBwuBglykwrtSogl0MymUHgboN4w
In this paper it goes into how these PAH degraders can be on both marine and terrestrial environments.
I agree things that humans see as toxic there could be some organism that thrives on that substance. The fact that micro-organisms are so abundant there are most likely thousands of organisms that we have not even discovered that could survive on alien planets.
Does the red pigment these bacteria produce indicate that they use this method of chemoautotrophy rather than photosynthesis just like most organisms that perform photosynthesis are green?
“The resulting slurry was collected in 150-ml serum bottles as thick suspensions, which were bubbled with N2 and crimp sealed.”
I was was wondering about the statement above. What do they mean when they say it was bottled as thick suspensions and bubbled with N2? Is this a method of storing the bacteria?
I don’t see anywhere that it says the mixture was higher or lower than the 9.3 pH. I believe what they were saying is that they were trying to achieve the same composition of the hot springs so in order to achieve this they used HCl to get it to the same ph. It didn’t make it more basic.
What could be the cause of the light incubated bacteria to be abruptly ended at 50 degrees C?
how do scientists know that something like anoxygenetic photosynthesis is an ancient process? Does this process leave some sort of impression or something of that nature?
Why is it important to mention that these organisms were obtained from a hyper saline environment?
The large errors associated with the sulfide-amended samples were likely due to the variable kinetics of formation and destruction of various thioarsenic intermediates which are soluble at this high pH (7, 20).
What does this statement above mean? What are variable kinetics that would affect the experiment?
unfortunately i read the end before the beginning..but had i read it accordingly, id have the same question as the others? this experiment seems to serve a minute purpose. we all want to know whats the outcome?
My first question is why did the samples need two months to be analyzed? Then next question is, only two samples? they’re very specific about the cleaning of the spatulas. is there another cleaning method that would disturb or perturb the results? In the Antarctic, it’s fairly cold, so now the question is raised, is there are bacteria that is specific to the area that would interact with diesel a certain way?
Why is it important to know about the degradation of aromatic molecules? What is the benefit for this experiment?
Is the soil in Japan deprived from vast amount of Oxygen? because the oxidase product is not isolated here, so does that mean the gene is not converted or is it because of the vast amount of arsenic in the area.?
im in agreement with Autumn here. I like the idea of balance here for the chemical version on respiration or photosynthesis. I am curious of which of the two states are more significant in this biogeochemical cycle.
if the primers are not perfect matches, does that not point to a mutation at some point? Because there should be a level of success otherwise. Mutated strands usually do not have the same capacity as its original.
i find it odd the color correlations here. Green colored water makes me think of swamps. I have been to the west coast and have seen the green waters there and they don’t seem to be a high temp. So the question here is, are the bacterias this color because this is the temperature they become dormant at? like is this color a resting stage for them?
is it dominated because of the relationship with Ectothiorhodospiracaea?
“Recent genetic analyses of the aromatic degradation pathways in PAH-degrading sphingomonads have increasingly revealed that most of the genes necessary for degrading an aromatic compound are scattered in several clusters and not organized in coordinately regulated operons”
The results of the last paper demonstrated similarities in the catabolic gene arrangement of sphingomonas sp. strain LB126 and Terrabacter sp. strain DBF63, the latter a gram-positive strain. In general, do gram-positive strains display a more coordinated arrangement of catabolic genes than gram-negative strains? Are these characteristics generally expected/revealed when studying various strains?
“Nevertheless, the biodegradation of the compound has been poorly studied, especially in terms of bacterial catabolic genes.”
Why is this the case? It would seem that the abundance of these molecules and their carcinogenic properties would merit more interest in understanding their degradation. Is this just a relatively new area of study in molecular biology? It would not seem so.
A general question but I am genuinely curious as it would seem bioremediation studies are rather popular.
“The DNA regions flanking the mini-Tn 5 insertion sites in mutants of strain A4 were cloned by digesting total DNA from the mutants with EcoRI, ligating them to EcoRI-digested pUC19, and transforming E. coli DH5 α.”
Why did they clone these particular regions in the mutants?
Regarding the RT-PCR results, am I to understand that they discovered what appears to an operon involved in PAH degradation but due to its inefficiency its questionable whether or not it would be worthy to investigate further? What would be a reason to further investigate the transcriptional units of such inefficient regions?
[The difference in the fold-increase of the mRNA level in the presence of acenaphthene between arhA3 (2.4-fold) and arhA1 (6.2-fold) remains to be explained, but it is possible that another, more inducible promoter(s) exists between these genes.]
Is there something else that would lead to this difference? Aside from the mentioned difference in stability of transcribed mRNA fragments.
[Nine genes in the genome of P73T were predicted to encode different ring-cleaving dioxygenases]
Are these predictions based off homology?
[However, the taxonomic distributions of the five plasmid proteomes of strain P73Twere different from that of the chromosome, suggesting the chromosome and the plasmids may have had potentially different origins.]
Is “potentially different origins” eluding to horizontal gene transfer. If not, could this statement still be indication of horizontal gene transfer?
[Notably, region B of the genome, which contained the PAH-degrading genes and were absent in another bacterium of Celeribacter, strain B30, was predicted to have been acquired via lateral gene transfer.]
“and were absent in another bacterium of Celeribacter”
What is this telling us? In context it seems to indicate horizontal gene transfer but I’m not sure how this absence in another strain would server as evidence.
I don’t think further investigation into these genes was done because the objective of the experiment does not require it.
The findings from gene P73_0346 (in region B) were probably sufficient for the publication.
I do agree that further research would reveal more and I believe that is the intention of this paper, to spark further investigation.
[The cDNA solution was diluted 10-fold in deionized water before PCR amplification.]
Why do they dilute the cDNA so much before amplifying by real-time PCR?
“These observations suggested that the genes involved in the initial oxygenation of acenaphthene did not function in strains AG2-45, AG2-48 and AG3-15.”
Is this implying that the genes did function in the AG3-69 strain? It would seem they did not according to prior sentences. I’m not sure I understand their conclusion here and how it was obtained.
[We postulated that ORF10 and ORF12 encoded an intrinsic ferredoxin and ferredoxin reductase for ArhA1A2 and tentatively designated them arhA3 and arhA4, respectively.]
Is this statement supported solely by the shared homology with the ThnA3 and ThnA4 genes in strain TFA? Is there support for this statement beyond this shared homology?
In the grand scheme of things, is the intention of molecular biologists to develop a strain capable of degrading PAHs in the ocean on a commercial level? This would appear to be the case but how practical or logical is this considering what is currently known about PAH degradation?
[Metabolic pathways have been proposed in many degraders; for example, Mycobacterium vanbaalenii PYR-1 (2, 4), Mycobacterium sp. JS14 (5), Sinorhizobium sp. C4 (6), Sphingomonas sp. LB126 (7), Rhodococcus spp.(8), Pasteurella sp. IFA (9), Staphylococcus sp. PN/Y (10), Burkholderia fungorum LB400 (11) and Pseudomonas sp. PP2 (12), but rarely in marine bacteria.]
Is there a specific reason for the scarcity of information on marine bacteria or is it simply due to a lack of studies?
I’d be interested in a comparison of results from similar experiments dealing with bacteria species extracted from deep sea sediment of various oceans/regions.
Based on their description of the degradation pathway of fluoranthene in paragraph 33 I would assume that the experiment did reveal genes for PAH degradation. At the least certain genes were assumed to be involved in PAH degradation based on prior research and homology.
[When this gene was disrupted by deletion with a kan cassette inserted]
Why was a kan cassette used for the disruption experiment? Can you elaborate on what this is and how it functions to disrupt the P73_0346 gene.
[The organization of gene clusters involved in PAHs catabolism of strains P73T and SL003B-26A1 were almost identical, with only a minor gene rearrangement]
Could this be viewed as evidence for horizontal gene transfer?
The use of indole as a color indicator to physically identify a certain strain of bacteria is fascinating. I have worked with agar plates that contains a specific antibiotic to sort out strains of bacteria; however, I never knew about the reactions of the indole and the dioxygenase enzymes turning indigo as a way of identification.
The isolation of Rhodococcus for the degradation of organic materials is very useful. Though it is great for environmental clean up of crude oil, what is the possibility of these bacteria strains causing harm to the surrounding living organisms? Is there a process to retrieve or eliminate them once the organic materials are depleted?
The use of the mineral salt medium is very common for microbial growth culture; however, why use salts? Salts cause a concentration gradient across the bacterial cellular wall and cause water from the bacteria to be drawn to create a balance; thus, shrinking the cells. Although there are bacteria that can tolerate the concentration, why is MSM widely used for most growth culture?
Incubating bacteria at 30C at 180rpm was a new lab technique to me. I was confused on why they needed to be shaken at 180rpm. I found out that if bacteria were left growing still, it would grow in layers, and the bacteria at the bottom layers would be depleted of oxygen and medium. Incubating at 180rpm would ensure that all the bacteria would growth evenly and efficiently to obtain maximum aeration. Some of you might already knew that, but I wanted to share something new that I learned.
Using PCR amplification as a mean for comparing nucleotide sequences and obtaining >80% identity with the databases is interesting. I did some research and the Rieske (2Fe-2S) protein sub-unit has a function to aid during electron transfer within the cytochrome b6f complex. This seems like a very important function and shows that the relevant matches in the database and the Rieske sequences within Rhodococcus have similar electron transfer proteins.
Is there any correlation or reasoning as to why Rhodococcus degraded only the aliphatic fraction of ILCO in both of the MSM and YMSM medium? With the incubation period being two weeks, what if the time was increased? Would that make any difference?
I believe the bacteria degraded only low percentages of ILCO because there should be a consideration of other organic matter present in the crude oil.
The utilization of three and four ring PAHs as a sole carbon and energy source for PAH-degrading bacteria can be very interesting when looking at it from organic point of view. Three and four carbon rings contain high torsional strain within the carbon bonds. Do these high energy bonds cause more reactions with the bacteria?
With the advanced technology of CRISPR-CAS, could it be possible to isolate the Rieske center region and incorporate it into a specific species of bacteria of choice that does not harbor the center originally?
I agree this this suggestion considering in the real world there would be several other chemicals and substrates present along with the bacteria.
The run off of organic materials in water systems because of agricultural use is very toxic to the environment.
The degradation of PAHs by marine bacteria could be very useful in the bioremediation of carcinogenic chemical run off in the environment.
With the use of kanamycin as an antibiotic for differentiation, is a certain strain of bacteria the goal of the experiment?
Was there a significant purpose as to why they used methanol instead of another compound to extract the products?
It is very interesting to see that the amino acid subunits of different species of bacteria can have the same type of cluster motifs.
The amino acid sequences of these species of bacteria are found to have a 5% divergence based on Clustal W. Does this indicate that the bacteria could contain the specific plasmid for the production of these amino acids?
This is a great point. It leads me to wonder if the most efficient bacteria from every environment could be bio-synthesized to have a dioxygenase gene from each bacteria for maximum efficiency in degrading PAHs.
This leads me to wonder if there could be a possible study that exhibit a comparison between the strains with the most efficiency in degrading PAHs.
I believe that the presence of the dioxygenase genes on the chromosome instead of plasmids suggest that the genes are essential for the growth of the cell. However, if bacteria cells share genetic DNA via conjugation with plasmids, are Cycloclasticus able to transfer these genes to other species?
[ This suggests that the electron transport proteins of the PAH dioxygenase system may be shared with other redox systems, possibly to maximize the catabolic potential while limiting its genetic burden.]
This suggestion leads me to believe that the transport proteins of this PAH dioxygenase system could act in a similar fashion much like a-ketoglutarate does in the way it gets generated in the kreb cycle and transported to another pathway for use.
The anaerobic cycling of arsenic is interesting. Further experimentation of this could be very helpful if it the genes for this degradation could be extracted and biologically transcribed by other species. This could lead to health related services when people are poisoned with arsenic.
That’s a great observation. I wonder if this ability was from their core genome or attained over time.
This in-depth description of temperatures and appearance in the site description tells me that is could be very important to identify the type of metabolic reactions that could be happening in the environment that the sample came from.
Great observation! Maybe the dark and low temperature reduces the slurry’s growth so that it could be preserved to use for several months? I’m not very sure either.
What a great point! It leads me to believe that they could essentially have a mutualistic relationship to benefit both parties in the community.
I wonder if bacteria could transform by using archaeal DNA and obtain this characteristic and increase their diversity?
I wonder why it took 40 hours of incubation to for the slurries to oxidize acetate. This seems like a random time frame.
This leads me to think about if this specific environment prevents organism from growing due to the clash between metabolic reactions and hypersaline environment, or do organisms just chooses not to live here?
Though these biofilms are found on the Paoha Islands, is it possible to introduce them to a new environment with similar characteristics and screen for activity or are these biofilms natively active?
The experimental incubations at similar temperatures as the hot springs suggest that they are adapted to that specific environment; however, it leads me to think that if we were to selectively grow them on different temperatures, it would eventually adapt and change genotypically.
Have they started using bioremediation yet to clean up these spills or are they still exploring options?
If scientists use bioremediation in a certain area and the microbes end up not working, what would be the effect? Would they just leave the bacteria in the water?
How are sampling sites determined? I noticed they do not give an exact location and the contents of the sediment is tested and discovered after the fact but do the researchers just go to random sediments and test the soil?
As the researchers determine which microbes grow best on different sources, how would they then be released into the areas where bioremediation is needed? I.e. would they just release a mixture of microbes into different areas in order to degrade everything that might be there or would they release only the best microbes to degrade whatever substance is most present?
How do the hydrothermal plumes and hot springs produce petroleum hydrocarbons? Are these the same as what we know commonly as gasoline/oil?
Since there are microbes that naturally degrade oil, are we studying them to see if there are ways to make them degrade faster?
What is the importance of incubating the flasks in the dark?
How do the primers amplify rRNA genes and what exactly is meant by amplify?
What is meant by mineralization by the bacterial community?
So they concluded that Cycloclasticus will possibly be able to degrade PAH compounds but were not able to get definite results in this study? I just want to make sure I am understanding correctly. n
Is biomineralization almost like bioremediation in this case? Since too much Se can be toxic and it is left up to bacteria to mineralize it so it doesn’t get to elevated levels, it almost mirrors how bacteria degrade PAHs in my opinion if that is correct.
When they say that only 2 genes have been found in the two bacterial examples, does that mean that only those two bacterial strains are the ones mineralizing the Se in waters?
Why are they using an antibiotic to select for colonies containing the vector?
What is the importance of knowing the shape and size of the particles of selenium here?
Since the E. coli S17-1 rapidly reduced the Se(VI) concentrations, how would scientists use this to reduce pollution? would they use the cells or use the organism to put into polluted areas to reduce the amount of Se?
What caused the changed in the diameter of the Se particles between organisms?
What do they mean by cosmid clones?
What does it mean when its saying the Se particles were associated with the exterior cell membrane?
are chlorate and bromate pollutants? Since the bacteria can use the enzyme to reduce those but not the others, would that still make them useful in finding efficient degraders for pollutants?
Are the organisms they use in these studies able to be relocated into areas that need the selenate degraders for the pollutant or is the main focus of this study to find the families of bacteria that can degrade selenate?
This paragraph was discussing the isolates strain of ZX4 and its effectiveness for degrading phenanthrene. Because it talks about potential one used for biremediation of oil-contaminated enviroments could this be used to help clean up oil spills?
Why is it a good thing for this bacterium to be used to produce indigo?
How come the Pseudomonas putida scattered compared to the Sphingomonas which were clustered in the same group?
If the microorganisms are used to remove the oil spill, how do scientist know how much microorganisms need to be placed in the oil spill to remove the oil? What is the effect on other organisms with using the microorganisms?
Oil usually remains at the surface, however it does sink down, can the microorganisms reach to a certain point to continue eating the oil at different depths? Has there been more research on the effect of the Oil spill in the Persian Gulf to this day?
Using the Gas chromatography was a way to extracts any condensed by evaporation. Is this a final step of in the scientists methods to isolate the naphthalene from the oil?Why would the concentration of naphthalene be incubated for seven days? The gas chromatography will give the scientist a percentage of the naphthalene.
From moving to different flasks and being filter could that possible cause any cross- contamination while trying to isolate the naphthalene? Could there be any other types of growth in the flasks?
What made the “18 isolated stains that show adequate growth rate” grow faster than all the other strains that were from the marine samples? Also in the beginning the article, it tells where each strain has came from and what they correspond with. For example N1 is a algae strain.
What would cause the adding this concentration to dramatic decrease the growth? As the growth decreases, the strain N7 had its maximum growth, what would cause this strain to have its max growth compared to the other stains?
Was using different researchers to obtained concentration used to compare data with the other researchers. As using the multiple researchers, it showed the E24 is the direct relationship between emulsification activity. What is the significance of E24?
How can isolates be helpful in bioremediation of PAH- contaminated sites and oil spills preserve the ecosystem in a natural way? In other words, what are the steps to preserving the ecosystem with knowing this knowledge?
Will the phylogenetic trees be compared with different phylogenetic trees that are base on 16rDNA? How was the phenanthrene-degrading bacteria strain being isolated?
Why will the scientist use meta-cleavage to catalyzed the conversion of catehols? And what would the scientists find with using the meta-cleavage method?
How can the isolated strain be identified by partial sequence of the 16S rRNA? How come they are using a European database?
Why was the medium suspended in the sterilized water? How would turbidity effect the medium?
What is ZX4? Why was the sample in the flask inoculated with a strain ZX4?
Looking at the table it should that the standard deviation of the ZX4 is going down. What is the reason for the PAH to be decreasing in 14 days?
Why was the lowering of C23O activity observed when ZX4 was growing? Was it because it was using the glucose and using the carbon source on the plates as energy?
Why were they unable to obtain authentic PCR products for arsenite oxidase genes?
If these bacteria were found in a hot spring, would most of them be chemolithoauotrophs if there is a lack of sunlight and use carbon dioxide for its source of energy?
Why were the slurries stored in the dark? Are they kept in the dark because then they could be viable for later experiments?
Is the methods that is being used (e.g BLAST) the same technique used in lab? How did the clones failed to show significant similarity?
In figure 1, it is showing how the samples shift from light to dark, but how come at about 70 hours the As (III) dies, then start having responses around 120 hours?
Why was the arsenate was completely reduced in the dark-incubated was samples?
In figure 4, why did the 2 mM sulfide sample that was incubated under an atmosphere of 100% H2 die off at about 70 hours?
How come the activity was 5- folds higher than the comparable dark- incubate ones? Could it be because of photosynthesis and the acetate prefer the light.
How could a primer are not suited for the environment? Also what is the alternative explanation?
I think it is interesting that the Paoha Island biofilms are a glimpse into the Earth’s distant past.
If the bacteria degrade these harmful compounds, such as the benzene rings, and then continue to be in the water, will the by-products created through degradation have a harmful affect on lifeforms as well? How long does this method have to be used before we can be sure of any side effects of this system?
Not only would environmental conditions impose selection pressure, but that combining with the uptake of different chemical components could potentially cause mutations within the bacteria used for degradation. How can this be monitored to keep a watch on any potentially mutations that could become harmful and decrease water quality?
Assuming that there is a notable mutation rate within the bacteria during incubation period, the PCR data could be skewed and cause a misread of the rRNA as a different bacteria that is not of importance to this experiment. Could satellite markers have been used in conjunction with PCR to spot possible small mutations?
If the time frame of this experiment were adjusted in either direction, would we expect to see a consistent rate of fixation or is there a peak efficiency time frame? At what point does the degradation become ineffective, either over time or during certain concentrations?
Would the growth of the various strains together create a competition over resources? Is it possible that the less productive strains would preform better in the absence of high preforming strains?
What differences in the strains cause this ability to handle different concentrations? Is this the product of mutations in the bacteria? Were these mutations already present or could they have occurred more rapidly though lab exposure?
With the bacteria coming from such a harsh environment, it seems difficult to appropriately replicate the extreame conditions in a lab setting. Will this affect the amount of oxidation/reduction taking place in the sample?
With the ability to transition between environments that have different oxygen concentration, could these extremophiles be some “missing link” ?
This sample is taken from a hot spring. Are there other instances of bacteria using toxic materials in their metabolism is less extreame conditions?
So instead of draining the waste in the environment can they collect them and treat them with microbes and then dump it outside? If those harmful compounds are abundant in the environment, how much amount will affect the mammals?
PAH bioremediation breaks the compound in CO2 and H2O than the level of CO2 will increase in water. Will this change affect the organisms in water, or there are no organisms because of polluted water?
Degrading PAHs using microbes is easy cheap, but will the overcrowding of that microbes at certain area harm the other creatures around them? Even before throwing the waste out, it can be treated with microbes and then let it out.
The phylogeny tree is always useful to understand the bacteria, as we get to learn which is similar to which and to draw the conclusion on the specific matter. To learn PAHs and their pathway of degrading will help us to understand how some plastic and metal cannot be degraded.
The colonies grew on the diluted plate would be light and when grown on fresh media will they grow faster than diluted as more resources are available to them?
was there any difference in growth when incubated in 28 degree C than in 20 degree C?
which carbon aromatic compounds were easily absorbed by them?
So with the help od content of G and C content of DNA we can find the Species and Genere of the microbes? what if the moles are same but have different placing order?
They were not able to obtain PCR product for arsenite oxidase genes, but instead of what they are assuming, they might be using a different gene or enzyme to reach the last step? Or they can even be the totally different process which is yet to be discovered?
As the reduction of As(V) and oxidation of As(III) can happen in both oxic and anoxic condition, they actually donot need CO2 or O2, and they might be the oldest living creature or one of earliest living things on earth?
Arsenic despite being toxic, which property of it makes it safe in respiration?
When does this change happen between one cycle to other?
why was the slurry bubbled with N2 gas? did they already know before that the bacteria might be using N2 gas?
why there was the linkage of arsenic biotransformations to chemoheterotrophic process?
the range was wide of As(V) because the bond between proton was weak.
Why did they chose this lake? why chose arsenite, and not methane for the studies?
There must be some sort of limitation on what we can or cannot do to protect Antarctica from human pollution. Guideline must be put in place to ensure that tourist only leave footprints and nothing else. Excessive mining of fossil fuel shouldn’t be allowed to preserve natural resources.
As I’m reading through the first few paragraph of this article, I notice this type of experiment is what I am learning right now. From the type of bacteria to the agar media. Moving on to creating primer to preform PCR. By using primer 27F and 1492R, the purpose was to make a copy of the 16s gene from 27th basepair to 1492nd basepair? In genetic lab, we called them by the name of the gene, which mean the primer F and R are from the same gene.
Is the primer 27F and 1492R from the same gene? From what I understand, PCR is basically making many copy of a segment of a gene. In this case the gene is 16s, is that mean they are making a copy from 27th bp to 1492nd bp?
I think PCR is rather common nowadays. We are actually preparing for a PCR in genetic lab right now. My group is using the gene name unc_27 in C.elegan.
Why did they conduct an incubation at 4 degree? That is very low, and why did they jump straight to 21 degree?
How did they figure out that there was cross feeding going on?
I am also interested to see how they managed to gather these microbes.
What is diagenesis?
What exactly do they mean by slurries? Is that just the mixture of the biofilm and rest of the ingredients needed to perform the experiment?
So the opposite reaction occurs in light vs dark conditions when there is no oxygen present, but when there is oxygen present that is not the case?
Maybe it has something to do with their enzymes not tolerating that high of a temperature.
How does oil contamination alter carbon/nitrogen levels?
I’m very interested in the human microbiome and all of the roles bacteria play in/ on our bodies. I’m sure the earth’s microbiome plays a huge role as well.
Is bioremediation of oil spillage more effective in cold weather environments? Why do they specify “cold weather environments?”
I’m not sure if they have found anything having to do with what you suggested but we are finding out ways in which the human microbiome is beneficial to human health. For example, bacteria that make up the “estrobolome” help women (and maybe men too.. idk) get rid of excess estrogen from their body. I’ve also read about certain microbes possibly protecting against skin cancer. (I’m not sure if that has been proven. It might just be a theory). Your gut microbiome has a good bit to do with your immune system as well.
Is stable isotope probing the best way to achieve their goal?
What is push coring?
What is porewater?
Why did they exclude the vector sequences?
What are singleton sequences?
What is pyrosequencing?
I find it interesting that arsenic is so toxic to us but helpful when it comes to helping bacteria derive energy.
Do you think their PCR attempt failed because of error or because these bacteria have different arsenic metabolizing genes?
What is crimp sealing?
I had forgotten what RFLPs were. I had to google it but I do remember hearing about restriction fragment length polymorphisms.
Can cooccurrence of As(III) oxidation + As(V) occur in oxygen rich environments by other bacteria?
Why was hydrogen added to the washed samples?
How did the 6 strains generate more phenanthrane fluorescence and why were they thrown out for it?
I would definitely assume that the bacteria adapted to the diesel contaminated soil, especially since other bacteria were killed off by it.. When humans take antibiotics, many have issues with yeast overgrowth that wouldn’t have had the chance to grow otherwise. The decimation of normal gut flora allows other microbes to grow.
The bioaugmentation was very interseting to read. Additionally, with advancing development or straightforwardly embeddings regions with contamination corrupting microscopic organisms, what might be the effect on the encompassing environment if this process was left unattended?
What was the idea behind the extraction repeating three times? Also, did the excitation-emission spectra make the chemometric computation increase or decrease?
Can iron through siderophores be effective in natural conditions?
How is bacteria used to benefit the iron that is being scavenged?
I did my research on CARD-FISH and flow cytometry because I wanted to know exactly what their role was. CARD-FISH can be used for phylogenetic staining of microorganisms in many environments, while flow cytometry is used to enable rapid analysis of significant number of cells at single cell wall. How does flow cytometry count cells?
[This process can be achieved by promoting the growth of endogenous metabolizing bacteria in contaminated sites (biostimulation) or by directly seeding contaminated sites with pollutant-degrading bacteria (bioaugmentation). ]
Is the bioaugmentation process similar to the process that was used cleaning the Deepwater Horizon oil spill in 2010? Also, with promoting growth or directly inserting areas with pollutant-degrading bacteria, what would be the impact on the surrounding ecosystem if this process was left uncontrolled?
[Oil contamination can generate detrimental changes in soil properties, including modifications in maximum surface temperature, pH, and carbon and nitrogen levels (Aislabie et al., 2004). ]
I googled the changes in temperature changes in Antartica, and found out that it is experiencing some of the most rapid warming on Earth. In the last 50 years, Antartica’s temperature has increased over 5 degrees Fahrenheit. Is this due to the reshaped microbial structures?
[Environmental bacterial isolates, as well as the control strains Escherichia coli BW25113 and Pseudomonas aeruginosa PAO1, were routinely grown in R2A and M9 minimal media. ]
Why would R2A and M9 minimal media be the choice of mediums for this research? Isn’t there much better mediums for E. coli, such as Luria Bertani broth?
[After 90 min incubation at room temperature, buffer contained in the syringe was recovered, serially diluted, and plated to estimate CFU/mL.]
With the buffer being incubated at room temperature, what would be the effect of adding a bit of heat. Would the 90 minute incubation drop?
[Samples were taken from the surface soil horizon (0–10 cm) from four sites exposed to diesel fuel, and four non diesel-exposed control sites,]
With all the sample taken from a soil horizon spanning between 0-10 cm shows that these samples are not from a particularly high horizon. Soils with higher horizon are exposed to more organic matter, and less human disturbance. Would taking a nutrient richer sample show a better perspective of how diesel fuel is altering the soil’s ecology?
I find it so interesting that a place like Antartica has a great diversity of bacteria. With such harsh climatic conditions, how is this possible? No trees, shrubs, and very few flowering plants have the ability to survive this ecosystem. Wouldn’t bacteria diversity decrease as the climatic severity increases?
[ Leave a comment on paragraph 4 0 Varied processes have been described to enhance the ability of bacteria to metabolize PAHs, including the formation of biofilms,]
It is amazing to learn all the ways, formation of biofilms help bacteria. I know bio films can physically protect bacteria from antibiotics and disinfectants, but this process takes resources and energy. With such harsh conditions in Antarctic regions; do these biofilms have a change in ability to metabolize PAHs compared to the ones in high nutrient ecosystems?
[ Leave a comment on paragraph 8 4 In environments harboring multi-species bacterial communities, siderophores are known to play roles in competition for iron]
Siderophores have both cooperation and competition interactions in their environment. To high of iron concentration can promote algae growth which can block off sun light and throw off normal feeding processes. My question is, why do siderophores have roles in competition? What is the advantage of siderophores sequestering available iron?
I researched that vibrio concentrations in oysters can be 10-fold higher than the surrounding water. Most shells fishes are great sources for iron. I was wondering could there be a correlation with siderophore aerobactin in vibrio and high concentration of vibrio within oysters?
[ The V. fischeri ES114 genome was scanned for siderophore biosynthetic genes using AntiSMASH with relaxed strictness]
I had to google how AntiSMASH worked for scanning siderophore biosynthetic genes, and found out that it allows rapid genome-wide identification, and analysis of secondary metabolite biosynthesis gene clusters in bacteria. With this being such a broad genome scanning, is there a larger chance of mistake?
[Unless otherwise indicated, erythromycin, chloramphenicol, kanamycin, ampicillin, and polymyxin B were added to final concentrations of 5 μg mL−1, 10 μg mL−1, 100 μg mL−1, 100 μg mL−1, and 50 μg mL−1, respectively. ]
I understand that these antibiotics were added to test the different resistance genes within the genome. My only question is, why are they added at such different concentrations? My guess is that the strength is huge factor when comparing the different antibiotics.
[following boiling or filtration through a 10,000 MWCO membrane, suggesting that the inhibitor is a small molecule ]
The molecular weight cutoff (MWCO) is in important term for the membrane filtration, describing the pore size or rejection ability of the membrane. Typical nanofiltration is anywhere between 200 and 2000 MWCO, so a 10000 MWCO is evident to an extremely small molecule.
[ It was curious that growth inhibition of V. harveyi occurred only when culture fluids were obtained from V. fischeri ES114 grown in minimal marine medium but not in rich medium]
This is a very interesting outcome because for most situations the rich medium would yield the better growth in colonies. Does this show evidence of the advantages of siderophores when iron is low in the environment?
[ Public goods producing bacteria are vulnerable to cheaters, bacteria that benefit from using public goods without paying the metabolic cost of producing them]
I understand how these cheaters have a competitive advantage over public good producing bacteria ,but is there a correlation between densities of both in the environment? If the advantage is strong, why is there a need for the cell to keep these genes that have a metabolic cost to make? I was thinking that for there to be cheaters, there has to be public good producing bacteria close by, and in higher densities.
[. Aerobactin production by V. fischeri ES114 could be especially relevant during colonization of its symbiotic host.]
I would imagine aerobactin production is a crucial part in colonizing a new symbiotic host. For reasons such as colonizing in a time of the year or after long draughts of rain where the iron supply is even lower than normal. Also, if the symbiotic host lives in a deeper part of the ocean where iron supplies are naturally lower than the ocean’s surface.
[Nitrogen losses from these oxygen minimum zones (OMZs) are estimated to account for 30–50% of total nitrogen loss from the oceans (9, 10)]
I always believed that the oxygen abundance would be similar in all areas of the sea. After reading about OMZs in the oceans, is there a correlation of water depth and oxygen abundance in the sea?
[A 20- to 40-m-thick suboxic transitional zone, characterized by low oxygen (<5 μM) and undetectable sulfide, persists throughout the basin between the surface oxic layer and the sulfidic anoxic deep water (≥100 m) (12, 13).]
I was wondering, with sea temperatures rising due to global warming, is this possible reason for the thick subtoxic zone in the Black Sea?
[. 15NO2 − production was measured in the same anoxic 15N incubation vials as in the anammox rate measurements, but 15NO2 − was analyzed as N2 after a two-step reduction by acidified sodium iodide and then by copper at 650°C]
Why was copper used in the reduction steps? Aren’t metals like magnesium and iron a more common metal found in the sea?
I did some reading on CARD-FISH to see the differences between in and FISH. CARD-FISH is a specialized form of FISH focusing on aquatic habitats with small, slow growing, and starving bacteria. It allows hybridized cells that are usually below detection limits to be signaled.
These graphs show distribution of inorganic nitrogen, sulfide, light transmission, and reduced manganese at different water depth, but focusing on the suboxic level. I gathered that from graph A, nitrate was at its highest level at about 82 m. In graph B the O2 levels drop significantly when entering the suboxic level dropping lower than 1 micrometer. Graph C shows that MnOx was at its optimum level in the suboxic zone.
This maximum likelihood tree shows likely hood of a phylogeny evolving are evolving from, and from this tree shows many clades that amoA genes were expressed at 80 m.
[Clearly, the production of 15N15N in 15NH4 + incubations was a result of anammox being linked to nitrifiers when no other NO2 − was readily available for anammox]
I thought the 30N2 isotope could be a product of anammox-nitrification coupling. Does denitrification also occur because if not shouldn’t there be a nitrite product in graph a from figure 2?
If we can use genomic analysis to identify which genes were transferred, could we also determine what bacteria the genes originated from?
This statement, has me curious about what else might be in the ocean that could help with the PAH issue that has yet to be fully researched.
What type of environmental conditions is required for PAH to strive in and is there an environmental factor that can prevent contamination?
Are non-sporulating aerobic rhodococci the most common found contributor to biodegradation because they are not giving all their energy to offspring production or is it something else?
Throughout most of the materials and methods I noticed that a multiple number of processes were incubated at 30 degrees Celsius combined with being centrifuged at 180 rpm for a week to two weeks. What was the reason for why they had to wait so long for each experiment before continuing and was there not a faster way for them to do these?
Even though the amplified PCR products were obtained and registered in a database, if the same soil bacteria sample were to be subjected to another experiment would the sequences be changed because bacterial DNA could be altered through conjugation or transduction?
I’ve seen multiple times throughout this research paper that everything that was extracted was analyzed by GC-FID. Was this the only instrument that they were able to use to get the results that they desired or was there another way to get the results but they didn’t have the equipment for it.
I noticed that the one gram soil sample was subcultured at a 1:10 ratio followed by a 1:50 ratio that was used for the 16 day period? Why was the ratio so low for the first initial step and then was stepped up to a 1:50 ratio for the rest of the experiment? What made it so necessary to have to use such a big ratio? to carry out the rest of the experiment?
For this table, when the researchers were determining what to use for the best relevant match, was there a set percent identity (nucleotide) that they were looking specifically for or was it just based on what matches were the highest that they could find?
What was in the medium that was supplemented by YE that made Nap not able to degrade? Was there a certain characteristic that it possessed to inhibit the degradation of NAP?
I was wondering if this bacterial strain screening process is still used today? I’ve tried to look it up on google but, I did not find any type of information correlating to that particular process.
When they were testing the addition of YE in mediums did they try different amounts of YE in each set of mediums or was there a generalized amount of YE that they would use for each Rhodococcus sp. CMGCZ sample.
I find it very interesting that the Cycloclasticus strains are able to to degrade substituted and substituted PAHs. I’m very curious about the extent of how efficiently they can degrade compared to Rhodococcus.
Are there specific species of marine organisms that can transfer the toxic PAHs to humans?
I agree with your statement. With the algae bloom in peak during the summer in the Gulf coast, research for the degradation of toxic chemicals would be helpful in retaining the marine biome.
What specific guidelines were used to determine the purity and the chemicals used when working on this experiment?
I agree, its a great learning tool to be able to learn about the techniques and processes in person as we conduct in person in the lab.
For the ONR7a medium is it better to have it enriched with naphthalene or phenanthrene or does that not matter?
I noticed that when the beta subunit was compared to PhnA2 it stated that it had a relatively high sequence identity of 52%. Is this considered to be high because the sequence identity is unable to get much higher than that or is it high because its over 50%?
I was thinking the same thing when viewing this figure, and to make matters worse I don’t understand why orf7 is going the opposite way as opposed to all the other open reading frames.
Someone can correct me if I’m wrong, but I’m pretty sure that the purpose of the flanking region is a region that contains the promoter and also may contain enhancers and or protein binding sites.
Madeline, I am too very interested in knowing if further studies will be conducted for the efficiency of other dioxygenase genes. I also wonder that if they are to find other dioxygenase genes if they will be much more efficient than the known genes or if they will not be as good.
Jessica, I was wondering the same thing and if so will it have more of a positive effect or negative effect.
Nolan, I am very interested as well to see how this study will further help the understand of the evolution of arsenic from the anoxic conditions compared to oxic conditions.
It is very impressive to see that there are organisms in this world that are able to withstand environments that would kill anything else when exposed to the conditions. Could these Organisms give rise to something that could be very beneficial to survival or could they just end up being a dead end road.
A periphyton to my knowledge is a microecosystem that’s composed of a complex matrix that is embedded with autotrophic and heterotrophic organisms
Taylor, I’m wondering the same thing. I have done extensive googling and I have yet to find another gene that codes for aerobic As(III) oxidation, but I also could be looking in the wrong place as well.
Alexandria, I was wondering the same thing, because I would think that neighboring lakes would contain atleast dissolved sulfide and ammonia.
Derria, I was wondering the same thing but my best answer for it would be that maybe they experimented with a various amount of temperatures and found that the set temperature that they kept them at is what worked best for them.
Spencer, I agree that you would expect to see more archaea in this type of environment, and I also agree with you that for once while reading a paper like this that I actually feel as if I have an understanding of what is going on.
Sarah, I was thinking that the reaction was able to proceed at a much faster rate because it was done within aerobic conditions, because correct me if I’m wrong but I thought that anaerobic respiration was much quicker due to the lack of excessive ATP that is produced, but I guess that it would only be quicker if it was done within anaerobic conditions.
Julie, I agree with you, when i first saw the figures in the results section it did not make much sense to me but when I read what was being discussed it helped a lot with the understanding of what was going on.
That would be amazing because we would be able to relate to the process and methods due to it being parallel to our lectures.
Was the PAH-degrading bacteria investigated after the BP oil spill?
Although the location had insufficient nutrients and high hydrocarbon pollution, what can be done today to attempt to isolate bacteria from the initial pollution site?
This was done to see if the isolated strain was the same as what they began with in the experiment.
How does the temperature of the bacteria at the sites chosen vary from the temperature at the site of the oil spill?
In Figure 2 it shows cocci and bacilli bacteria on nutrient agar plates.
The figure helps to visualize and makes more sense of what is going on in the experiment. As well as show why some isolates grew well on some agar plates than others.
Why hasn’t deep sea sediments been investigated or researched more with the growth of production of oil in deeper water provinces?
How does the vent fluids affect or change the sediment around the surface?
In this steps photos of sampling sites display hydrothermal features and sampling sites without hydrothermal features.
In this step the potential strain were mineralized and grown separately in a liquid medium. Following this the cell’s biomass was washed several times prior to use to ensure sterilization.
Why was the core brought to the surface then cooled?
What exactly is Clustal_X?
In this section it explains how one must be careful to ensure the experiment yields results that are interpretable.
What would be the oxidized intermediate that caused the yellowish coloration??
How was the makeup of the dominant DNA different from the others?
How does the gene increase and the disappearance correlate ?
Of the two cosmic clones, why was the cosmic clone pECL1e chosen for further characterization versus the other?
In this figure, it shows for pECL32 the transcriptional direction is to the left, gene location to be in fnr and ogt and that it is positive for selenate reductase activity.
How is there so much known about how microorganisms affect and use Selenium but not about the biomineralization of it ?
How do genes control selenium reduction?
For the selenium reduction experiments, why were the cells initially grown in sealed Pyrex bottles containing LB broth and not on an agar plate?
After it was determine that S17-1 would be the E coli host what does it tell us about its makeup ?
I was curious about the Antarctic treaty that apparently prevents “the introduction of foreign organisms into the Antarctic continent,” so I looked up the details. The treaty was set in place in 1961 and holds Antarctic aside as a scientific preserve and allowing freedom for scientific investigation on its land. It does not allow for any military presence or any kind of weaponry testing anywhere on the continent. The treaty allows for Antarctica to be the one place on Earth that is uninfluenced and untouched by human interaction. It is essentially a natural and pure laboratory for the world to use carefully.
I was surprised to learn that they heavily relied on diesel oil in Antarctica as well. It seems like microbial bioremediation would be a good solution temporarily, but overtime I’m afraid with the increasing issue of global climate change that it would not be enough to keep up with the growing contamination and pollution. Especially as human activity only continues to grow. I am under the impression only so much can be done with the limitations Antarctica is under as the article states “bioaugmentation can only be implemented by the use of native microbes.”
I was curious about the other methods use for quantifying chemotaxis other than the modified capillary assay mentioned in the article.I found that other methods use bacterial concentrations that are too high to inhibit chemoeffector consumption or require conditions that make quantifying too burdensome. The technique of modified capillary assay for quantifying chemotaxis is actually an inexpensive procedure and can be apparently used with volatile or semivolatile chemicals.
Non-polar compounds were chosen to extract from the culture media. Extraction was repeated three times. I wondered why in this particular step of phenanthrene quantification, only non-polar compounds were extracted instead of polar as well and why specifically three times for extraction?
S. xenophagum D43FB and P. guineae E43FB were both shown to have the ability to adhere to phenanthrene crystals, however only D43FB was chosen to further study by scanning electron microscopy. Is there any particular reason why this isolate was chosen over E43FB even though they were both shown to exhibit strong and intermediate staining?
[Finally, although D43FB failed to show chemotactic responses, several genes encoding components of the flagellar system were found, suggesting that this strain has the potential to swim and respond to attractants in its environments, under the proper gene inducing conditions. ]
It is apparent that D43FB has strong potential as a good PAH-degrading bacteria candidate for a bioremediation, I however am not fully convinced it is truly the best candidate. The next question or set of experimentation for the researchers is to see if the strain can perform well under the conditions found in Antartica. My concern is that it will not perform as expected when the carefully crafted conditions created by the laboratory dissipate when they are tested in all actuality.
[While S. xenophagum D43FB was unable to produce biosurfactants and did not exhibit chemotactic responses, this strain exhibited the ability to form robust biofilms in vitro and was able to adhere directly to phenanthrene crystals, as shown by SEM microscopy, suggesting this bacterial isolate can tightly interact with phenanthrene when using it as its main carbon source.]
As previously mentioned in the article, a bias was found in the experiment because the strains were grown under standard laboratory conditions. While it is great the researchers found D43FB was able to form bioflims, it also occurred in vitro meaning an experiment performed in a test tube or laboratory dish. I would infer that this does not guarantee that S. xenophagum will always possess this ability especially when not completed inside of laboratory conditions, but in the actual environment.
Like many of my other classmates, I too was curious about the different methods of bioremediation. I found one article that lists the advantages and disadvantages of using In situ to ex-situ bioremediation. In situ is typically less costly because equipment is not required to unearth the contaminated soil and it is cleaner overall because it does not circulate dust and pollutants back into the surrounding area. One of the disadvantages is the extended length of time for decontamination. The three main strategies that fall under In situ are Biostimulation, Bioaugmentation, and Bioattenuation.
[The remarkable finding that atmospherically-transported Saharan dust enables proliferation of vibrio bacteria by delivering dissolved iron to surface marine environments further demonstrates the exquisite scarcity of iron in the ocean ]
I too was confused on whether iron benefits or impedes bacteria, and thought looking up the interesting finding of the Saharan dust delivery on the ocean surface would help clear up some confusion. The article I found had a team travel to the Florida and Barbados to assess the levels of Vibrio growth in ocean surface water during natural Saharan dust events. They found in just 24 hours of exposure, Vibrio background level went from 1 percent to almost 20 percent. They also say vibrio has the ability to quickly respond to nutrient plumes at a faster rate than microalgae which could reveal that Vibrio could play a major role in being intermediaries in the biogeochemical cycling of iron.
[Surprisingly, in our initial attempt to characterize the co-culture system, we found that V. fischeri produces and releases an inhibitor that prevents the growth of V. harveyi.]
In the previous paragraph, the researchers said that they chose Vibrio harveyi and Vibrio fischeri because they are known to occur together in nature. My question is why then does the problem arise with V. fischeri releasing an inhibitor and preventing V. harveyi from growing under standard laboratory conditions?
[V. fischeri ES114 makes almost no bioluminescence under laboratory conditions so the presence of any residual cells did not contribute to the bioluminescence reading.]
I thought this was interesting and wanted to know why exactly it would make almost no bioluminescence under laboratory conditions? I researched what could be the cause and found that when in low density like open marine sea, the luminescent genes are turned off.
[Plasmids were transformed into E. coli by electroporation using a Bio-Rad Micro Pulser. ]
I was curious about the process of electroporation so I decided to research it and found that it is the physical transfection method that uses an electrical pulse to create temporary pores in cell membranes in order to increase permeability of the cell membrane. This allows chemicals, drugs, and electrode arrays to be introduced into the cell. Interestingly, it can also be used in tumor treatment and gene therapy since it does a great job of introducing foreign genes into tissue culture cells.
[Thus, the difference in siderophore production between V. fischeri ES114 and V. fischeri MJ11 does not stem from differences in transcriptional regulation, rather, the difference apparently arises at the protein level, perhaps due to differences in post-transcriptional regulation, biosynthetic enzymatic activity, or protein stability.]
I understand that the researchers tested the efficiency of biosynthetic enzymes in V. fischeri ES114 versus in V. fischeri MJ11 by overexpressing the iucABCD operon from each strain in a V. fischeri ES114 ΔiucABCD mutant. The exact cause of this variety in siderophore production is not exactly specified by this test. I was wondering if they were other tests that the researchers could run to determine between all of the possibilities, which one is the determinant?
[qRT-PCR confirmed that luxT was transcribed from the overexpression vector in E. coli as there was a 40-fold increase in luxT transcript levels in the strain supplied with the arabinose inducer compared to the isogenic uninduced strain ]
I was unsure what the exact meaning of “isogenic” was and found that it refers to organisms having the same or closely related genotypes. Concerning the rest of this paragraph, I see that it was essential for the researchers to perform some ground work by testing the limits of LuxT and how it will contribute in comparing and contrasting vibrios species competition.
[First, possession of AerE makes an aerobactin cheater immune to cytoplasmic aerobactin toxicity. We say this based on our assessment of the growth defect displayed by the V. fischeri ES114 ΔaerE mutant (Fig. S10A). Second, possession of AerE may enable aerobactin recycling by cheater vibrios, fostering higher overall iron acquisition and, in turn, a superior growth advantage during competitive situations.]
Just because a growth defect was observed with the V. fischeri ES114 ΔaerE mutant during the experiment, is it fair to conclude that any and all possession of AerE will make an aerobactin cheater immune to cytoplasmic aerobactin toxicity?
[Moreover, both vibrio species produce a variety of public goods including extracellular proteases, chitinases, and QS autoinducers, all of which can be monitored in real time.]
I was curious as to what exactly a”public good” meant exactly in this context. I did a google search and found an article defining it as “biological public goods are broadly shared within an ecosystem and readily available. They appear to be widespread and may have played important roles in the history of life on Earth.” They are essential factors that have played an important role in the evolution of life like merging of genomes, protein domains, etc.
[Nevertheless, the identity and abundance of the responsible nitrifiers, or any coupling between nitrification and nitrogen losses, remain poorly documented.]
Are these couplings poorly documented because it is difficult to record data at such deep depths of the ocean?
I wanted to know more about Crenarchaeota so I looked it up and found that it is known for having microbial species with the highest growth temperatures of any organisms. This is why they have been isolated from and able to sustain in deep-sea marine environments.
[Anammox bacteria was verified by CARD-FISH with the probe BS820 (15), but strong background fluorescence precluded accurate enumeration and qPCR was used for quantification. Total microbial abundance was measured by flow cytometry (57).]
I was curious to see what exactly was flow cytometry and found that it is a technique used to detect and measure the physical and chemical characteristics of cells. Is this method a default when the population of cells are hard to observe?
The CTD system detects and measures the temperature, pressure, and conductivity of seawater. These factors all affect what kind of microbes are present in the various depths of the ocean. I researched what were the main differences between the two types and found that pumpcast enables high resolution water sampling along vertical profiles, while rosette cannot sufficiently detect small vertical structures in the vertical distribution of trace metals.
These various graphs for figure 1 confirm a couple of things. In the introduction the researchers state that the suboxic zone can be characterized by low oxygen and unobservable sulfide. The anammox bacteria levels are high at this zone which also confirms the notion that nitrogen loss via amammox occurs here. Figure 1B and 1D both verify these statements respectively.
These graphs correlate to the figure 1 graphs regarding the nitrification and anammox levels in their respective zones. In figure 2A, it can be seen that mRNA grows exponentially well in the suboxic zone with the ammonia-oxidizing crenarchaea. In figure 2B, we see poor growth from mRNA and BAOB cells. In figure 2C, mRNA did not follow a strict growth pattern, varying quite a bit. YAOB cells grew the most in the suboxic zones.
[These observations may imply that in such suboxic settings, these crenarchaea were not using their nitrifying capabilities much but some other energy-acquisition pathways.]
What might be some of the specific reasons that ammonia-oxidizing crenarchaea were not able to utilize their nitrifying capabilities in the suboxic zone?
[Despite the barely detectable gene abundance, strong amoA expression by AOB was detected within the nitrification zone (Fig. 2). γAOB amoA expression, in particular, was up to nearly 3-fold greater than that of crenarchaea. ]
I find it interesting how the gene abundance of amoA was so low, it still however expresses so strongly in the nitrification zones. Although it was strongly expressed, is it possible that the low gene abundance is the reason why it did not measure up to its predicted net nitrification rate previously set by the researchers?
I find it interesting that the present species Rhodococcus sp. CMGCZ is able to degrade higher concentrations of Fla than the previous species of Rhodococcus. I’m wondering what caused the change between the homology of each species and what changes to the homology makes the present species different from previous species?
Since the present species of Rhodococcus is able to degrade high concentrations of Fla would that change the cycling of the pollutants or interfere with the function of marine and terrestrial ecosystems?
When I first read this paragraph the first thing that caught my eye was bead-beating, and so I decide to look it up to figure out what it is. I believe this is a lab manual or some kind of manual, but in it the author states, “be a debating is accomplished by rapidly agitating a sample with a grinding medium (bead or balls) in a bead beater (device that shakes the homogenization vessel).” I hope this helps anyone else that was just as confused as I was when I read the words “bead-beating.”
https://opsdiagnostics.com/notes/ranpri/OPSD_Bead_Beating_Primer_2014%20v1.pdf
I believe the purpose of washing the cultures before they are resuspended, is to make sure that they are sterilized before the next step in the procedure. I’m wondering what exactly the cultures are being washed with, unless I over looked it earlier in the paper.
What does finding the sequences and putting them in a phylogenetic tree do for the experiment? Does it just make it easier for the observer to see the difference sequences or is it used for something else?
Madeline I was thinking the same thing! I’m wondering what is exactly causing the Fla to be completely degraded by YE, but Nap cannot be degraded at all and Phe is slightly degrading with the presence of YE.
Why did the degradation of Fla decrease with the increase of its concentration? Is it because there wasn’t enough time to degrade a higher concentration or is there something else involved?
I find it interesting that this chart shows what each sequence is most closely related to, but also how closely they are related by percentage if I am understanding the chart correctly.
Is it the different concentrations of YE in different mediums that is changing the degradation of Fla? Or is it also because they are changing the concentration of of Fla?
Why does an increase in Fla concentration have an inversely proportionally effect on degradation?
In the previous paper we just read about how PAHs are pollutants in water, so I believe that the difference between coastal and terrestrial is that they come from different sources. But I am interested in finding out what else makes them different.
I agree that it would be very interesting if the same genes are responsible for degrading both marine and terrestrial PAHs, but if they have different sources would the same genes be able to degrade both?
If PAHs can be transferred to humans through the consumption of seafood can they be transferred to humans through other ways?
What exactly does the M9 medium do? It contains ampicillin, thiamine, and glucose, but they do not explain what exactly they are using the medium for. Like lab this week certain tests are used to test for certain things, so what does the M9 medium test for?
I find it interesting that we have essentially done part of this in lab; where we put a culture on a plate and see if it has blue colonies. Its also interesting to see how the things we do in lab are used in different experiments and how we can relate what we have done to the experiments we read about.
Do both clones pH1a and pH1b both have the same 10.5-kb Sau3AI fragment? If so why only those two clones, if they got pH1a and pH1b from the genomic library wouldn’t all the clones they make from it have the same 10.5-kb Sau3AI fragment?
Why would the identity of the amino acid sequences be different if it is the same molecule? Are they different because they are using different alpha subunits or is it because of something else?
I find it interesting that they have less than 44% of their amino acid sequence in common. I also find it interesting that alpha and beta subunits had similar trees.
It’s interesting that PhnC is involved in the upper and lower pathways of degrading naphthalene. If it is involved in the upper and lower pathways of degrading naphthalene does that mean that it is more efficient in degrading it?
Christy, I was thinking the same thing. In an earlier paragraph it talks about how PhnA1 has alpha and beta subunits, so I’m wondering what effect of having one or the other or even both would have on their ability to degrade hydrocarbons.
I find it interesting that the oxyanions arsenate and arsenite are able to be a redox couple. And that the redox reaction that can help other organisms gain energy for growth. I find it interesting that Arsenic has different uses, besides it well known use of being toxic.
If As(III) and As(V) are found in periphyton, does that mean that the redox reaction that is occurring between them is helping the periphyton grow? That is what I’m getting from paragraph two if I am understanding this correctly.
I believe that the primers could have been the problem for not being able to obtain an authentic PCR. But could it have also been because of the enzymes that they used? After obtaining the PCR in lab and talking to Dr. Ni Chadhain she talked about how some enzymes could work for some genomes and not for others.
If find it interesting that they combined the PCR products. What was the purpose of that, did it help them create the clone library?
What exactly is the biofilm used for? Is it helping with extracting the DNA? If so how does it help with extracting it?
I find it interesting that As(III) and As(V) had opposite effects when being introduced to the dark regimen and the light regimen. Is this happening because it is showing how one can be an electron donor and one can be an electron acceptor and vice versa?
I agree that it is interesting that they came from the same biofilm, but have different temperature ranges. I’m wondering if this has an affect on their incubation experiments with being in the dark and light regimen.
After observing figure 4 its interesting that As(V) and As (III) have graphs that look very similar. As(V) and As(III) start out at different points but each graph looks similar to the other but inverted. I believe this is showing how one can be the electron donor and the other can be the electron acceptor in a redox reaction.
I find it interesting that even though acetate is important in anaerobic processes is was not found to have any stimulation on As(V). I’m wondering what could be that cause of this and why does this happen.
Jessica, I was wondering the same thing. I feel like they could have done the incubation experiments to make sure that what they thought was correct, but I’m not exactly sure.
While it is very interesting that organic and inorganic compounds can increase PAH degradation, how would this practically look in day-to-day life? Would we add these compounds to potentially contaminated foods? And how would we limit respiratory uptake? As someone mentioned above, I can’t help but wonder what the consequences would be in the environment.
Can it be assumed that Rhodococcus sp. is the better option when compared to the original strain of Rhodococcus? And between Rhodococcus sp. and inorganic nutrients such as nitrogen, which would be a more practical way of degrading PAHs in the environment?
After knowing the results of the study, I wonder what effect nitrogen or phosphorus would have on degradation of PAHs if used in conjunction with CMGCZ?
What is the benefit to incubating the sample at 180rpm? What would happen if the sample was not continuously mixed for 2 weeks and just kept still?
Since degradation was also examined in YMSM plates to determine the effects of yeast extract on the degradation potential of CMGCZ, I’m curious to know if MSM or YMSM plates supported higher rates of degradation. I think it will be interesting to see if the YE had any effect on CMGCZ.
After using this similar technique in lab to grow black and blue colonies, I now see why they let the plates incubate for 2 weeks instead of 48 hours.
Why was CMGCZ unable to degrade Nap in YMSM, but had a higher rate of degradation for Phe and completely degraded Fla? Is there a nutrient supplement that would support all 3 PAHs instead of just 2?
When looking at Fig. 2, the degradation of Fla in YMSM from days 1-3 appears to resemble the Fla degradation in MSM from days 5-7. It seems that YE in the YMSM plates shortens the time that it takes for Fla to be degraded. While I think the effects of YE on Fla degradation is interesting, I wonder what the effects of other inorganic and organic nutrients would be in comparison to YE.
Originally when I thought about the effect that YE had on CMGCZ’s ability to degrade Fla, I was only focusing on the fact that it completely degraded Fla and ignored the fact that it was unable to degrade Nap at all. I can now see where CMGCZ’s inability to degrade Nap would leave that PAH in the environment, whereas unenhanced CMGCZ at least has the ability to degrade a small amount of Nap.
Before Rhodococcus sp. CMGCZ is used for bioremediation purposes, I think more research needs to be done on it. More research could lead to a strain of Rhodococcus sp. CMGCZ that degrades Nap and Phe at higher levels instead of just Fla. Also, the effects that this would have on the environment, whether good or bad, are still unknown. Despite the need for more research, I think CMGCZ could prove to be a great option when it comes to degrading PAHs in the environment.
I think it would make sense for marine bacteria to do a better job at degrading PAHs in marine environments than terrestrial bacteria. I would like to know how Rhodococcus compares to the two terrestrial isolates mentioned here.
This study focuses on different PAHs than the previous study which makes me wonder if there is a difference in types of PAHs found in marine environments versus terrestrial environments?
I wonder what the effectiveness of the transformed E. coli was versus the original Cycloclasticus when it comes to PAH degradation.
Maybe I’m missing something, but what is the benefit to adding antibiotics? I thought this study was also focusing on PAH degrading capabilities of a bacterial strain.
Are there any disadvantages to forming the monohydroxylated forms instead of the cis-dihydrodiol forms? It says that the cis-dihydrodiol forms are the typical products, but doesn’t say if the monohydroxylated forms cause any problems.
Even though the PhnA1 amino acid sequence exhibits a high percentage of identity with the amino acid sequences of other bacterial strains, I wonder which strain is the most effective at degrading PAHs. I also think its interesting that PhnA1 exhibited 51-62% sequence identity while other alpha subunits exhibited levels of sequence identity lower than 45%.
Since phnA1 and phnA2 alone did not result in an active dioxygenase, but did show dioxygenase activity when coexpressed with phnA3 and phnA4, does this mean there is a link between dioxygenases and ferredoxin/ferredoxin reductase?
What is the significance of the alpha subunit and beta subunit sequences falling outside of the major cluster?
I wonder if the different location for PAH degradation genes has anything to do with the difference between terrestrial and marine bacteria?
Just because PhnA was unable to convert anthracene and monocyclic aromatic hydrocarbons, could there be other genes in Cycloclasticus that could convert those hydrocarbons?
I wonder if these bacteria/archaea could provide any benefits to other arsenic rich environments besides hot springs, and could they use any other toxins as sources of energy?
I’m interested to know what role these arsenic metabolic activities might have had in the Archean earth, and how they have adjusted to all of the environmental changes since then.
What is the purpose in testing both dark-incubated and light-incubated tubes?
Is it common for clones not to show any similarity with anything from the GenBank database? And why were the sequences that didn’t show similarity with anything excluded from further analyses?
I think its interesting that there were more types of Archaea that were found than Bacteria, but at the same time I would almost expect this due to the harsh environment they were found in.
I found it interesting that the temperature ranges differ based on if oxidation or reduction is occurring and whether or not they are exposed to light or dark. Why does the presence of light affect the temperature ranges for metabolic activities?
I think its really interesting that a new type of aerobic As(III)-oxidizing microbe could have been found in this system! I wish they had used an additional primer in this experiment so we would know if it was more likely that the lack of results came from using the wrong primer, or its a new microbe.
I think it would be interesting to determine which organisms are responsible for the dark As(V) reduction.
I think its really cool to think about how the processes that are being studied have been occurring for billions of years, even though the earth has changed so much since then.
Even though the Archaea are not involved in the arsenic cycling, I wonder what their role is in that environment? I understand that they are there due to high salt concentration, but what is it that they are doing?
I know that even the most remote areas of the earth are still affected by the actions of humans, but I did not know the extent of our presence in Antarctica. I also did not previously consider the effects on microbial communities. Usually when people think about environmentalism they automatically think about larger, well-known, and charismatic organisms such as penguins.
I appreciate the fact that the proposed solution to this problem is using the natural abilities of the bacteria that already exist within this environment. It’s almost like the earth is trying to fix itself and self-heal, and we just need to help the process become more efficient. I also wanted to mention I think that it’s really neat we are doing essentially the same experiement in lab as what is descibed here.
Both PCR and Sanger sequencing are extremely important experimental methods that changed science and allow us to do research like what we’re reading about in this paper. However, I did some reading to refresh myself on how these processes work, and I found that Sanger sequencing is actually considered more expensive and inefficient compared to next generation sequencing. This makes me question why they chose the Sanger method and if there are specific reasons as to why Sanger sequencing was used in this case.
I think the biosurfactant production is important to measure here because sufactants will help to break down the PAHs present in diesel oil, including phenanthrene.
The three highest phenanthrene-metabolizing strains were all taken from diesel contaminated soils. I wonder if this is because the bacteria there have been adapting to the presence of the diesel, and how long did it take for the bacteria to be able to metabolize diesel this efficiently?
I was surprised to read that the bacteria are actually only psychrotolerant. I assumed they would be psychrophylic.
Could these processes be used in warmer environments too?
It’s comforting to know that they identified a few different strains that are known to be useful for degrading PAHs, but that there are possibly more that just haven’t been found yet.
I’ve never heard of “marine oil snow” before but I think the term is a clever way of decribing how oil can fall to the sea floor.
I like that both this paper and the previous one are exploring unique environments like Antarctica and the deep sea floor.
Why do the oil droplets show up as red?
I recognize the bacterial primers used here as the same ones we just used in lab for our own 16S rRNA experiment.
What makes them hard to cultivate and how could we possibly solve the issue?
Why were some treated with acid again?
I did some quick googling to get an idea of what vector sequences, chimeras, and singleton sequences are. I vaguely remember learning about chimeras before, but vector and singleton sequences were new to me.
I remember learning about that experiment!
I understand that C13 isotopes are carbons with an extra neutron, but how are they accessed for use in an experiment like this? Are they collected somehow or can you create them?
Why are there fewer reports in (HMW) PAH- degrading microorganisms? Is it because they are harder to work with/to find? It is interesting to hear how these bacterium can degrade such a damaging neurotoxin. However, do these bacterium cause any negative effects to the environments?
It is really interesting to find out how this process works! I have done similar techniques in other classes but never really understood how the reactions worked.
What effects do chloroform/methanol solvent produce on the bacteria? Is it a growth accelerant?
Will we be doing this at all in our labs? I think this is an amazing tool to help advance research and upload possible new sequences.
What is the purpose of removing the 37.8% of Fla on the third day? Why not just wait till the eighth day and remove them all at once?
If there is such a minimal percentage in the difference between the MSM and YMSM would have it been easier to exclude one of them in the beginning? Or would this be different depending on the bacteria inside the sample?
I’m curious if the different strains of degraders are used for different applications or if they all can be reproduced in the environmental remediation, pharmaceutical, and chemical industries.
I wonder if Rhodococcus is practical to use/obtain. I always hear of different types of bacteria but I don’t understand how easy it is to replication/obtain. Would it provide adverse effects to use it frequently?
I am curious as to how old this paper is. Did this paper influence any further research, experimentations, or practices of this species in the field?
If the product of Rhodococcus sp. CMGCZ is not reported in the ARHDs would this make the results only subject to this specific one? Have they tried replicating this experiment to see if they got similar results with different dioxygenases?
Upon reading this paragraph I was reminded of how in our last paper they experimented to see how these PAH degraders were able to digest PAHs within the Rieske center. I wonder if they will perform similar experiments here.
I am excited to see these degradation genes broken down and analyzed. It will be interesting to see what information they gather from this analysis.
I like that they are also looking at the physical applications of PAH degraders and which environments specific species would better thrive in.
Will we have access to the tables in this study before we begin our papers? Also, I tried to look up Luria-Bertani agar plates but did not find results as to what their purpose may be in this experiment.
What is the purpose of taking genomic DNA and partially digesting it with an endonuclease wouldn’t this skew the results?
In relation to restriction mapping since these clones had the same fragments does that mean they are a closely related species?
What difference does the presence of the cluster provide?
Would the fact that it falls out of the major cluster affect the degradation potential for this gene?
This paragraph made me think back to the first paper. I wonder what the amount of dioxygenase α-subunit genes were reported for that species of bacterium.
If both As(III) and As(V) can be reduced/oxidized in both oxic and anoxic conditions then why would they not experiment in both conditions?
While interesting that they were not able to obtain the PCR products I was curious if these biofilms are the experiment or if they were made as a precursor to what they did.
How did they know that the fermentative alkalithermophile Anarobranca californiensis was isolated from that area of the hot springs? Was it tested before and they are assume nothing has hanged?
When removing biofilm why did they change from using the sterile toothbrush to the sterile spatula? Does this make any difference?
Why do we not see more organisms present in total? I understand that it is an extreme environment but I always assumed there would be thousands of different types in any given environment or is it typical for a few (6 in this case) to dominate an area?
Since these incubations were cultured in a sealed bottle would have been beneficial to the experiment to remove the waste products? or would it have contaminated the results?
I’m curious if other saltwater microorganisms in the ocean exhibit a lot of similar characteristics or if this lake is substantially higher in salt content compared to the ocean thus the same characteristics or environmental pressures wouldn’t be there.
Why would the temperature range vary? I expected both the As(III) oxidation and the anaerobic As(v) reduction to be productive at approximately the same temperatures. Is it simply the light-driven have adapted to addition temperature due to heat produced by the sun?
What was the purpose of this incubation experiment? It seems they started off knowing the answer so was it just to verify?
If it was determined that acetate amendment was not present in the As(V) reduction “implying that chemoheterotrophy did not drive reduction” then what would be responsible for the drive of As(v) reduction?
I’ve noticed in this paragraph and the one before that they are consistently monitoring and regulating the temperature of the samples. Is this simply standard practice or are PAH-degraders sensitive to temperature changes? If so is this sensitivity the reason there are not many variants of PAH degraders?
I am struggling to understand why they are performing these steps. I understand that they were isolating the PAH degraders but I was thinking that they had already done so in the earlier steps.
It is said in this paragraph that these degraders are from different classes. I wonder what characteristics are observed to be different between them
I am confused why they say that P.centronellolis does not have any reports of PAH degradation. However i’m the previous paragraph they mention that it showed good results in phenanthrene degradation.
I was unsure of what culture turbidity was. After some searching I found that it a way of measuring the amount of particles suspended in a liquid by using light
I tried looking online to find out what the clear space around the strain indicates but am still unclear. I think it has something to do with lysis
Is the movement of the petrochemicals from the vents In the wild a necessary replication in the experiment?
I wonder what the difference is for pah degraders in the ocean and those elsewhere.
I was unsure if falcon tubes were a specialized type of use. Upon looking it up I found they were simply clear tubes that allowed you to see the sample.
I do not understand E.coli’s use in this part of the experiment. Wouldn’t this just blur the result?
I am curious if these strains are also common above ground/areas other than the ocean
I just want to make sure i’m understanding this section correctly… Since dna-Sip is a delicate process they preformed three variations of incorporating carbon into the biomass of the obtained sample degraders. This was for comparison of the results of carbon degradation and pah degradation that were run simultaneously?
I noticed that they are using the neighbor-joining method again in this paper. Is this the most popular technique?
So is this paragraph saying that in a experiment separate to the one in the previous paragraph Cycloclasticus was found in other parts of the sample core?
I like that this part of the experiment allowed them to confirm that these strains are able to grow on and degrade PAHs
So for this experiment are they looking into the aerobic or anaerobic Se-resistant microbes?
Since E.coli has the gene that is required for selenate reduction then the researches will not be able to use it as a control as they did in the last experience that, correct?
After some research, I saw that a cosmic cloning vector is used to clone large fragments of DNA to grow that DNA as a virus.
I don’t understand why it was important to look at the XAFS of Se atoms in this experiment. Is this before or after degradation?
What does pECL32 verify that the loss of Se(VI) reductive activity was due to disruption of Fnr genes?
I wonder if they continued this study to identify the other operons that contain the FNR binding site i’m the promoter region.
Does this mean that they have identified the only pathway of selenite reductive activity or only one of them?
Why do the fused rings and absence of terminal surfaces in PAHs provide resistance to biodegradation? Is this simply because rings tend to be more stable compounds.
I would be interested in seeing a compare and contrast between the Nakheel beach region and Kuwait and Iranian coasts. I wonder how drastically these PAH degraders if at all in the Arabian Gulf region.
Does bioremediation present any ethical concern or issues? For example, the creation and use of genetically modified organisms because of the potential risks that may appear from altering a genetic code.
Are there any downsides to using bioremediation? Such as possible spread of these bacteria to other areas? How could you control where these bacteria go? Could they possible have a negative effect on an area which does not require bioremediation?
The CTAB extraction method is known to be one of the best methods for DNA extraction. Are there any downsides to using this method? For example, contaminants from plant tissues?
What is the purpose of keeping the flasks on a shaker throughout the incubation? What would the results be if they were kept stationary?
In reference to the N7 strain, what characteristic could be present to allow it to have maxiumum growth at 400ppm? That high of a concentration of NP created a drastic decrease in growth in all other strains but N7 exhibited maximum growth at these toxic levels. Why?
It is evident that the N7 strain has the highest growth in comparison to the other strains throughout the whole process. Regardless of the concentration, N7 is always showing more growth. But overall, the figure resembles the pattern of a bell curve, meaning there is a steady increase in growth – then a peak of all strain growth around 400ppm NP – then they all show a drastic decrease above that concentration.
In what ways does horizontal gene transfer interfere with the phylogeny of the gene (xyLE) that encodes for catechol 2,3-dioxygenase?
What was the purpose of cloning the operon genes for the catechol meta-pathway and glutathione-S-transferase? Was this because they already knew those genes were useful in degradation and detoxification? Or that they wanted more present so they could further classify them?
Alignment of DNA sequences refers to the process of arranging these sequences to determine segments that may be similar to one another. This helps to further compare the strains of DNA and see which ones are similar as a result of structure, function, or evolution.
What was the purpose of transforming the DNA fragments into a plasmid then into E. coli? Was E. coli chosen in particular?
In the extension, or elongation, process of amplification is used to add a new DNA strand that is complementary to the template. This usually doubles the amount of DNA target sequences. Each time the extension/elongation step is complete there are more strands available to act as templates for the next round. Consequently, this leads to the exponential amplification of the target sequence.
Why did they choose to characterize this strain in particular out of multiple that were probably isolated? Did they have prior knowledge to this strain that determined it would be the best to further research?
Is there any significance to the strain being about to degrade via two different pathways? Salicylate and meta-cleavage pathway? Is is commonly found in PAH degrading strains?
Since there is no previous report on the meta-pathway gene of S. paucimobilis being able to degrade PAHs would this be the first discovery for this strain? Could this lead to the discovery of more bacteria capable of using this pathway to degrade PAHs?
What type of genetic modifications could they perform to improve PAH degradation? Would they be genetically modifying the strain to contain more meta-cleavage pathways?
The nitrifiers and denitrifiers exhibit a symbiotic relationship to one another. The process of one provides sustenance to the other, and vice versa.
This is probably the best microbiological evidence provided so far that suggests the possibility of life elsewhere in the universe.
Is the color appearance of the springs caused by the type of bacteria that inhabit it? Is the green appearance due to the cyanobacteria and red appearance due to the purple bacteria?
There is a big temperature gap between the storage of the the slurry sample and the biofilm for DNA analyses, why? The slurries were only needed to be kept at 5 degrees C and remained for several months. Where as the biofilm was storeed at -80 degrees C only until extraction.
That is correct. In the dark, anaerobic environment the Arsenate, which acts as the electron acceptor, needs an electron donor to be reduced to Arsenite. When no donors as present it cannot be reduced because no electrons to accept. Sulfide is a somewhat decent electron donor so it gets reduced a little bit. But H2 is the most favorable electron donor, to all of the Arsenate is fully reduced due to the abundance of H2.
Archaea are more diverse than Bacteria because Archaea are more similar to Eukaryotes.
Even though the range of temperature of the As(V) is wider than the range of As(III), both exhibit the same optimal temperatures.
The natural sulfide and H2 present act as electron donors for the As(V) (electron acceptor) to be reduced to As(III).
In these environments, there are many microbes present so this slight decrease in As(V) (or increase in reduction) was due to abiotic processes of other microbes that may be around.
If i had to make an educated guess, the term “in vivo” means occuring within/inside another organism and “de facto” means that something is actual, or in fact.
So, for them to use those terms it would mean they are saying it does “in fact or in reality” occur “inside the organism”.
It is important to research which bacteria will do best in what environment to ensure the best results in naphthalene degradation. Is there a way to lessen the presence of other microorganisms that might compete with naphthalene degrading bacteria? Or should we look for other options for PAH bioremediation by observing the way nature breaks down cyclic aromatic compounds in similar environments?
Even though PAH bioremediation is considered effective and benign, if large amounts of carbon dioxide are released during this process then the possible ramifications need to be considered. High levels of carbon dioxide are seen as a problem in the environment as a whole and the adverse effects within marine ecosystems have already been noted. If PAH bioremediation was used how would we remove the excess carbon dioxide from the environment? Or would we continue to contribute to the threat of marine ecosystems and marine life? Other alternatives to PAH bioremediation should be considered and preventing more harm to ecosystems should be our top priority.
How accurate of an estimate does the turbidity measurement provide? Is there another technique that would provide more accurate data but still be feasible to preform? Does the calibration curve ensure that naphthalene is fully removed? How do you determine a calibration curve and why is it necessary?
What nutrients do the ONR7 medium provide in this experiment? How do you determine which medium would be best to use? What kind of benefit does the rotary shaker provide? Would bacteria still grow without the use of a rotary shaker?
What is the reasoning behind using 16S rDNA gene sequences that are available in public databases for comparison? Is this because of the availability or because these databases are found to be the most accurate? What is bootstrap replicates and neighbor-joining analysis? Why is this the best way to infer tree topology? Would there be a better way to infer tree topology even if it took longer or was less convenient?
What is the major difference between N1 and N7 strains when compared to N16 and N18 strains? What makes these strains more productive in degrading naphthalene? What is the GC-FID method and why is it more feasible to use to analyze microbial growth when compared to other methods?
What kind of marine environments are best for isolation of naphthalene degrading bacterial strains? What makes some marine environments better than others?
Even though some of the genera can uptake crude oil, are there any consequences of it doing so? Does genera produce any harmful effects in the environment?
Is microbial transformation and degradation also thought to be the best method for removing other pollutants from the ecosystem? Which strains are the ones that can degrade PAHs completely and what are their limitations?
Why was there a need to clone the catechol meta-pathway operon genes and glutathione-S-transferase gene? And what was the reasoning behind thinking this strain would be good for performing bioremediation of PAH pollution?
How was 200r min-1 determined to be ideal for the culture? What is the procedure for determining it?
What is the point of disrupting with 99cycling of sonication for 3 seconds? What differences are identified with a molar absorption coefficient for CDNB and GSH?
Are the tests that were done typical for bacteria characterization? Are these the main tests that most use to identify bacteria? Are there any more tests that should be considered and why?
What is GST and what does it mean by revealing it was CDNB accepting type? How does GST increase give us information?
What is meta-cleavage and what does it involve? What does the clustering of strains mean as far as results?
Is the meta-pathway present in all bacteria able to degrade PAHs? Or does the meta-pathway make it more efficient? How would understanding the meta-pathway facilitate the improvement of PAHs degradation through genetic modification?
What is a diel cycle? What are other energy linkages between aerobes and anaerobes?
What is the reasoning behind not being able to find a PCR products? Could another microorganism have utilized the products produced?
What was the reasoning for bubbling with N2? Does the biofilm have to be placed on dry ice to preserve it? What would happen if dry ice was not used?
How are clone libraries constructed? Were groups containing one or two more clones selected because of time consideration? If not, then what was the reasoning behind it?
What is the reason that all aoxB clones failed to show similarity to anything in GenBank? Why is RFLP analysis the way that the clones are grouped? What makes this analysis the best?
What is the point in depositing the sequences in these different databases? Is this just to add the information for reference during other studies?
What was the point of incubating samples under atmosphere of 100%? And what was the purpose of amending with 2mM sulfide?
Is sulfide or H2 a better chemoautotrophic electron donor? And why was the sulfide condition not tested when looking at acetate assimilation?
Why would this cyclic phenomenon only show in oxic/anoxic experimental biomes? Isn’t it necessary even in completely anaerobic ecosystems? How does the cyclic cycle differ from that in anaerobic ecosystems?
Is it more likely that the primers are not suited for the environment or that there is a novel organism present? What ways would we determine what kind of mechanisms the novel organisms use for As(III) oxidation?
Through oil spill is very critical, I personally still believe that nuclear leak should be the most critical environmental pollution. in 2011, there is a case of a nuclear leak in Fukushima Daiichi, Japan. I heard that it will affect the coast and living organisms for over 50 years.
But this paragraph is to introduce bioremediation, I may not really put questions on the giving background.
I think the yeast plays the role of catalyst, it is used to speed up the reaction and they may want to set up the experiment to test PAH removal in different concentrations and bacterias. it is easy to lower concentration by adding solvent. The no substrate 100 ppm PAH is more likely a control group or starting material.
In figure 2, 2 samples are incubated at 21 degrees, but I think in the deep sea, the bacteria should habitat in a much colder environment like what material and methods page mentions 4 degrees but the method page also mentions 21 degrees. Is that mean those bacteria in the deep sea can grow at room temperature and function similarly in both temperatures? if so, can we find those hydrocarbon-degrading bacterial somewhere else like the crude oil on the surface of the sea?
I’m not sure how do they figure out which antibiotic s can be used in the transconjugants?
Why proving that fnr gene can solely active selenate reduction result in only fnr gene? Is it possible for ogt gene can also solely activate the selenate reduction? In figure 2, it also just shows the graph about proving the function of fnr gene, is there a technique issue for testing with only ogt gene?
I think it states that E.coli specifically contains selenate reductase because of the non-oxygen environment in the intestinal which does not need the oxygen-sensing transcription factors to detect the absence of oxygen and regulate selenate reductase activity like other cells with oxygen. in other words for aerobic cells, the selenate reduction needs a transcription factor to regulate. But I don’t understand how the hypothesize given the bacterium adapts from aerobic to anaerobic.
If it is a catabolic pathway it seems it would be more prominent around higher concentrations of PAHs. Could an increase of the concentration of microbial degraders offer information about the environment? Thinking more in marine environments. Or even soil? It may be easier to test for the actual PAHs.
The genetic acquisitions of Alteromonas sp. SN2 is that referring to horizontal gene transfer and is it possible to use genomic analysis to identify which genes have been transferred?
Maybe PAHs aren’t the sole source of carbon, or these microbes have the ability to degrade and not necessarily be completely dependent on them? Then the mutants could still be viable. I’ve started working with some arabidopsis mutants that have a disrupted continuum of cell wall, plasma membrane, and actin cytoskeleton leaving them struggling to produce much cell mass. They are pitiful compared to the wild type, but enough for our purposes.
P73T isn’t the only gene responsible for degradation then. Alis is right it would not survive in this environment or have metabolites for that matter. It’s interesting to see where this is going. How complex is the pathway and the mechanisms involved?
The first paper in the reference section was extremely useful. It is a mini review on PAHs and the basis for degradation by microbes.
The possible potential on P37T for use in oil spills is exciting. It is quite remarkable to learn about how many genes have been linked to HGT, and to consider how many more marine microbes possibly hold similar potentials.
This is making sense to me(hopefully). It would be like looking into a (arabidopsis) mutation of actin depolymerizing factors (ADF) and testing strains of ADF3, ADF4, and ADF5. Systematically testing slight variances to get at precise mechanisms of functions.
I’m very excited for our 16S rRNA gene comparisons in class. Personally I can find clarity in concepts more readily when performing tasks, or at least gaining a visual for the process in its entirety.
B30 seems to be a very good comparison against P73T. It makes a strong contrast for the genes in B and D in P73T.
I wonder if the novelty of the P73_0346 has anything to do with it being a marine bacteria. In the introduction it was stated that proposed metabolic pathways for degraders have been rarely seen in marine bacteria.
My understanding is that COG annotation is representative of speciation events, which in bacteria happen quite rapidly. So the database was constructed to show genetic differences between closely related species, I think it may highlight HGT?
This is why PAHs accumulate? They are hydrophobic therefore not readily susceptible to non-metabolic degradation. Making them “persistent organic compounds”(intro)
How do scientists determine which naphthalene-degrading bacteria to introduce into the environment? If these bacteria are not able to compete with other microorganisms in a certain environment, how do scientists determine what to do next for naphthalene degradation in that environment?
Were the time frames between each assessment of the growth curves of the isolates the same each time? Also, could a different assessment routine have shown better results?
What are the other aromatic compounds that the two-ring naphthalene is compared to in this paragraph? What is meant by “more easily?”
I recall the BP oil spill where so much oil was dumped into the ocean that the animals were coated and the coast wasn’t safe to swim in anymore. This is what sounds like the result of us beginning to travel and explore the Antarctic more will be. We see the major effects these events have on macro-organisms but pay very little attention to what happens to the microorganisms.
The more natural and protected we can keep Antarctica the better because there is a lot of potential for studying bacteria that has been frozen in the layers of ice there. Not only to mention the native species only found in Antarctica, the fact that only native microorganisms can be introduced is a relieving statement as dumping so many foreign organisms into the water would surely have an effect on the native species.
This is very cool to see that the experiments with the microorganisms being done here could possibly lead to a way to degrade oil in the future. This, if done correctly could lead to ways for us to help restore areas that have been damaged by oil spills.
Typically PCR is used for DNA replication but I believe that is only for a segment or certain strand. They were likely trying to dilute and isolate a certain colony of bacteria that could use a specified fuel source. Once they isolated the bacteria however, I do believe that PCR would have been a plausible option.
Were the bacteria collected with the syringe the ones that were to be isolated for further replication? That is what I had gathered from this as it seems that the bioattractant was used to separate the desired bacteria from the other bacterium.
I recall that as well, it seemed that the values were from 25-250 listed in the manual. I am unsure of this but I think that the video linked in the pdf said you could count up to 350 and that would be an acceptable number.
In this experiment the first time the bacteria replicated it took much longer to replicate, such as with a 48 hour lag phase, but in the next culture it didn’t take near as long. I am wondering why it took so long for the bacteria to replicate, the first time especially. The culture mediums would have had a large amount of nutrients that the bacteria could digest, and normally in rich media bacteria replicate rapidly.
That’s what it seems like from the previous posts. I believe it was in the introduction that the idea of dumping bacteria into the ocean to remove oil spills by bioremediation was suggested. I would imagine that the same idea or process would be used for this, though in this paragraph in the second sentence it states that Antarctica is a region that introducing forbidden material is forbidden so that may hinder the process or the research here could at least be used for alternate situations.
I am wondering id they will be able to use this research in Antarctica. In the second paragraph it was stated that foreign materials can’t be introduced into Antarctica, so does that mean this research is only for finding bacteria to be used in other situations?
That’s what it sounds like to me. I would imagine that the oil bunched together and got too dense to float on the water so it eventually descended to the ocean floor.
So would the bacteria that they are testing be autolithochemotrophs, and barothermophiles or baropsychrophiles?
Since the bacteria are psychrophiles would this be the highest temperature they could tolerate, and since bacteria grow better at their upper range of temperature tolerance then wouldn’t that mean this would theoretically be the best temperature for maximum bacterial growth?
They probably froze them so that the bacteria wouldn’t replicate and essentially become a culture broth. These bacteria are psychrophiles so they would likely excel in replicating if they didn’t completely freeze them.
I think that is likely the answer. The PHE is probably the most similar to PAH and there was likely a mutation allowing in the enzymes/proteins that allow for the degradation of PAH.
I understand that the acid inhibited bacteria are used as a control group, but why are they using acid controlled bacterium specifically. Wouldn’t they simply be dead bacteria?
I imagine so, unless this was the maximum temperature the bacteria could tolerate since bacteria tend to metabolize and grow more at their maximum tolerable temperatures.
The 13C model was the only one being observed in the study since the 12C and the 14C were just mainly for comparisons it seems. So if they weren’t being studied then I imagine they were only concerned about results from 13C.
It seems like the As(V) is the electron acceptor and As(III) is the electron donor since they are listed as a redox couple. It also seems like they have isolated chemoautotrophs as well as photoautotrophs meaning yes, they will use CO2 as carbon sources.
I would imagine that this was were they decided to do research on this bacteria in Japan or that they did in fact find that the soil in Japan had a higher concentration of Arsenic making it more hospitable to these bacteria.
I am slightly confused as to how they adjusted the pH to 9.3 using HCL as it is an acid. I would think adding an acid to a solution would make it more acidic rather than making it more basic.
That sounds to be likely what happened, either that or the research on these primers were conducted in conjunction with the research on the bacteria in the biofilms.
Wouldn’t the differences in the concentrations mainly be the result of these bacteria likely being photolithotrophs?
Does tis mean that these bacteria are functional or micro anaerobes since adding oxygen nor removing it effects the rate at which they degrade materials?
Yes acetate is the result of removing a proton from the alcohol group that is in the carboxylic acid group.
Would this mean that this is possibly a new form of lithotrophy that hasn’t been identified yet, and is likely specific to Arsenic at these oxidation states?
What makes the yeast enhance degradation? Could there be another co-substrate that could be added to enhance degradation where the PAH ppm doesn’t have to be lowered?
It is weird they found a unique PAH-biodegration pattern and never isolated it or investigated it. What could have caused this and why was it only in one location?
Does the acetone help to estimate bacterial growths? Does the oven temperature for Pyrene and phenanthrene help with degradation?
Was there a difference in the PAH when the oil spill originally happened from after they did PAH degradation? Did the PAH degradation make a major impact?
With the concentration of extractable PAHs in the sampling locations high, what is the normal range? Do the phosphorus and nitrogen levels have any correlation to why the PAH is high?
Is there a reason to why all the isolates had different colony morphologies but were still rod shaped and gram-negative. Would there be a difference if they were gram-positive versus being gram-negative.
I had the same question Jessica. I was confused as to why they mentioned it in previous paragraphs but went back on what they said.
Lundy that is a great question. Now that I have read what you have said I am also confused why they haven’t run more tests on P. citronellolis.
Why was there such a big difference in temperature between the core collected in November and December? Does the temperature difference have to do with the well developed Beggiatoa mat?
Hi Melanie! Great question I was wondering the same about what clearing zones where. I understand now that when adding the PAH it makes it cloudy and when it is removed it turned clear.
Hi Tessa I totally agree with you that its very interesting to see how this could go in so many other directions. It’s great to see how this work added to the knowledge we know about hydro-carbon degrading bacteria.
Hi Anna I was also confused on what effect new were suppose to be seeing since the PAH was removed. Thank you Dr. Chadhain for clearing up my confusion.
Hi Madelyn I was also confused as to why E. coli S17-1 was not able to reduce Se(VI), but E. coli S17-1 combined with cosmid clone E. coli pECL1e was able to reduce Se(VI)? Does this have something to do with being grown in the minimal salt medium?
Does the diameter difference between the two mean anything significant? What would have caused one to have a bigger diameter than the other?
Hey Cody! I totally agree with you that this experiment was to show the importance of the fnr gene
Hi Danielle! Thank you for explaining more about cosmic clones I now understand why they used them in the experiment.
Hi Melanie! I agree with you on the conclusion of the discussion. FNR is a potentially control for Selenate.
I have the same question I was confused on whether chlorate and bromate are pollutants.
Lundy that is a great question I was confused as well on how the hydrocarbons are recycled.
I am interested to see how many PAH-degrading bacteria they will find in the Guaymas Basin since deep-sea has not been seen often.
How has the mechanism not been understood better since this is causing biological effects on animal life?
I am interested in learning the mechanisms and direct cloning in the results.
Good question Jasmene I was wondering the same thing. Also did they add antibiotics to all mediums?
Noel that is such a great question I was wondering the same thing. Could another method be used instead of this one and give the same effects?
I think it’s extremely interesting to see how different bacterial species can degrade various toxic chemical groups in similar manners. I am definitely curious to see the next few sections on this paper to see how their experiment compares to that of the PAH degraders.
I was interested in reading exactly where PAHs come from and how they are released into the environment, and what I found was that these chemicals are found naturally in coals, tobacco, wood, and other materials that release PAHs when burned (sited below). The “harmful effects” listed in this paragraph also seems indicative of smoking cigarettes and second-hand smoke, which are both ways that PAHs may enter someone’s body.
(https://www.epa.gov/sites/production/files/2014-03/documents/pahs_factsheet_cdc_2013.pdf)
The efficiency of the catabolic processes of the rhodococci to transform these harmful PAHs into TCA cycle intermediates is extremely fascinating. It seems that these bacterium play a crucial role in degrading environmental contaminants for human health and also in the nitrogen and carbon cycles for maintenance of atmospheric conditions.
I’m noticing here that these researchers used MSM plate media, just as we did in Lab 2 to begin isolating our naphthalene degraders. The composition of MSM seems to be commonly used to enrich the growth of the PAH’s.
After performing the initial steps to isolate our naphthalene degrading bacteria in lab today, I am curious to start to see results and draw connections between our lab and what these researchers are finding. As someone stated before me, perhaps incubating for 2 weeks will have more of an effect than incubating for 48 hours – I believe we will be able to observe and record the differences seen among the time frames in our own lab work in the next few weeks.
The results of the degraded samples on the MSM (for Nap and Phe) are contrary to what I thought they would be. I drew a parallel between the researchers using MSM in this experiment and our class using MSM for our soil sample in lab to a significant contribution in the degradation of PAHs. It is also interesting to see how significant the degradation of Fla was on both MSM and YMSM plates compared to the other two samples.
This makes me curious about what our MSM and R2A plates in our own lab experiments would look like if we observed them over a period of 8-14 days, instead of 2-5 days. I checked my group’s plates 48 hours after the experiment and only had one plate with blue/black colonies. It will be interesting to view our plates on Tuesday and see if we can make any connection between our results and this experiment.
When I was reading the “Results” section on the efficiency of CMGCZ to degrade the different PAHs, I was very interested in reading that Fla (HMW) was degraded 100% after 8 days on the YMSM but Phe (also HMW) was not significantly degraded after 8 days. The report mentioned in the “Introduction” that there have not been many reports on degradation of HMW compounds, so I found it peculiar that a HMW compound used in this experiment was degraded entirely.
I wondered the same thing. One would think that a smaller, aromatic structure would be more easily degraded than a larger aromatic. In addition to there being such a striking difference between the Fla and Nap degradation, also throw in the comparison of the Phe degradation. While Phe is also a HMW compound, the degradation more closely resembled that of Nap (LMW) than Fla.
My interpretation of this experiment based on the “Materials and Methods” section was that the researchers were changing the concentrations of Fla in the medium and recording how efficiently the CMGCZ would degrade the various concentrations. This experiment was performed on both MSM and YMSM mediums.
As we read further into this paper, it will be neat to draw comparisons between the Cycloclasticus here and the Rhodocuccus bacteria from the last paper. It looks now as though degradation of petroleum is a major role of study of the bacteria is paper 2.
It is interesting that this paper places more of an emphasis on the effects that PAHs have on marine ecosystems, rather than terrestrial (i.e. human) ecosystems. Perhaps the researchers wanted to study effects on marine life that could possibly have an effect on terrestrial lives through seafood, water, and other possible ingested pollutants.
An expression vector is a plasmid or virus designed for gene expression in specific cells. In this experiment, I presume pPhnA and pPhnC are plasmids that will be inserted into dioxygenase genes.
I would think that a defined medium would be used to eliminate the number of variables affecting the growth of the bacterial species. If it were a complex media, it may be more difficult to isolate the colonies needed for the experiment.
I found by researching online that LB agar plates are a common complex media used in microbiology research. I presume that the purpose for using this kind of agar is to allow growth of multiple forms of like (E. Coli and the recombinant plasmids).
In lab 6 that we completed this week, we also used a type of endonuclease to digest the nuclei of our naphthalene degrading isolates. It is starting to become easier to draw connections between our lab work and the work performed in these research experiments!
It is not directly stated, but I am assuming the aromatic oxygenase gene is the gene responsible for degrading PAHs? The introduction of the paper does not specify a particular gene they are searching for, just that Cycloclasticus sp. should have degrading capabilities.
I am extremely puzzled by this figure. Each of these structures (A, B, C) are physical maps of genes on the plasmids – I understand that. However, I don’t understand how restriction sites play a role in the ORFs? I can not deduce why they are shown in the figure or their significance.
I am a little stumped on how this paragraph says “the order of [genes] was found to be quite different from that of analogous genes reported previously” as well as “A5 harbored no plasmid… localized on the chromosome.” Are the researchers implying that they found something new with their research, or that previous accounts were incorrect regarding the location of these genes? Or are they simply pointing out that they found different results?
I’m noticing the term “cluster” being brought up quite a few times throughout the Discussion. What exactly does this mean? For example, “it falls outside the major cluster”; I’m not sure how to interpret this.
That’s a very good point! I never considered the idea of a PAH degrading bacteria to be categorized as an extremophile, but for some reason an arsenic-degrading bacteria does seem like it might fit that category.
I’m just wondering here, why did they want to place the samples on an artificial hot spring medium? I would think they here is when they would want to determine specific composition and abilities of their bacteria, so why wouldn’t they use a selective media?
So, I’m just making an educated guess here, the radioassay is what the researchers are using to determine the strength of the arsenic-degrading genes in the bacterium? Because they already know that these specific genes are present based on the genomic library construction, this portion of the experiment is now testing those genes?
This table is interesting because it shows more percent identity with species of archaea than with bacteria. I guess because archaea are generally less-widely studied, it surprises me that the majority of their matches were with these species.
It is so interesting to see that there are opposing effects that As(III) and As(V) had in the dark versus in the light. I would think that this is happening as a sort of oxidation-reduction coupling, where both reactions must take place to keep the cycle going.
Reading this discussion is definitely helping me connect a lot of the dots we saw in the figures in the Results section. Where this paragraph states “which implied the involvement of physiologically different anaerobes” seems to make more sense to me in regards to the differences in the dark and light degradation of arsenic seen in the last section.
It is interesting here that they did not find the results about acetate and chemoheterotrophy that they were expecting to find. Are they stating that the strains of interest capable of degrading arsenic are not chemoheterotrophs? Or are they simply saying that acetate is not an electron source for their metabolic functions?
I think this is really interesting too. When studying for this last exam, I remember reading something about how it’s easy to study the 16S rRNA gene because it is “highly conserved and universally present” among various types of organisms. Pretty neat!
Lameace, I too am curious to know how they would go about re-testing this since the first set of primers was unsuccessful. Would another type of primer work? Or would it be better to use a transcriptomic approach, such as a gene chip microarray? Is there a benefit to using one over the other?
Since PAH exposure occurs via respiratory uptake and some of their effects seem to deal with the respiratory system, does that mean they generally occur from instances where smoke enters the air, like smoking cigarettes or burning stuff?
So after reading the paper in its entirety, I’m getting the vibe that Rhodococcus is a key player in degrading PAHs, and that it is one of the few bacteria that can degrade HMW PAHs. Are HMW PAHs worse than LMW PAHs?
Hey Alexandria! I was wondering the same thing you were and did a little research to find that Rhodococcus can degrade a lot of things! Some interesting things I found that it could degrade was estrogens and alcohols.
By using the different types of media, are the researchers testing to see which offers a better source of growth for the PAHs? Also, why were two separate stock solutions of 1% Fla made? What purpose does spraying the agar with Fla made in acetone have?
I was wondering the same thing Lameace! I know that PCR is used to create copies of the DNA sequence of the target molecule but I’ve never given it any thought as to whether or not another gel could be used. Like you said, maybe a certain gel is used for a certain purpose and the researchers used the 0.8% Seaplague GTG agarose for a certain function.
I am thinking the same thing you are Lameace. I would think the washing of the bacteria would help eliminate any potential contaminants to the sample and allow for the culture to be pure.
Compared to R. erythropolis in MSM, the R. erythropolis in YMSM degraded at a faster rate in the first few days but seemed to catch up in a significant drop off by day seven. It even states in paragraph 7 that there was a delay of degradation in the MSM as opposed to the YMSM, which was delayed for the first 24 hours. I’m wondering what media, then, would be most beneficial to use given their seemingly different rates of degradation but overall similar effects?
So this may be a dumb question but I had no idea that biologists or molecular microbiology used aspects of organic chemistry, like proton NMR or IR to analyze compounds! I could also be reading the information and chart wrong, and it is some other kind of analysis, but either way, it looks really cool!
Based on the cluster information from the Rehmann et al. (2001) study, it seems Rhodococcus sp. CMGCZ falls into the second set. Although it was able to use other PAHs besides Fla, it definitely favored that PAH in particular.
With the finding of the Rieske [Fe2-S2] center, do you think it could potentially serve as a better degrader of Nap, Phe, Fla, and ILCO or limit Rhodococcus sp. CMGCZ’s already established success? Personally I would love to see further analysis into it.
This introduction is very similar to the first paper we summarized. It seems like the main difference is in the testing of a separate bacterium from Rhodococcus.
So this study’s primary focus is more so on marine PAH degraders than terrestrial ones? I wonder what aspects of marine PAHs make them unique from PAHs found on land.
I never thought of it like that until now! You’re totally right though, Tara, I wonder how much harm we are actually doing to our ecosystems in the name of maintaining it and allowing controlled growth? The grilled meat releasing PAHs is sad too, looks like I’ll be baking my chicken from now on.
I’ve always wondered what motivates a researcher to choose the type of analysis and tools of measurement they use because I can imagine that there are many different ways. How do they know that the one they chose will be the best fit for their intended study.
Is it a requirement for all researchers to report their nucleotide sequence data into these genetic libraries or is it a sort of professional courtesy?
Even after watching the video describing the cloning process and going over it in class today, this process is still very confusing to me. I am starting to understand bits and pieces of the process however which is good progress in my eyes.
What is the point of examining the substrate range for the ARHD? Does it allow the researchers to gauge the versatility of it?
Is there any significance to the fact that PhnC activities were similar among all catechol compounds? Also, does 3-Methylcatechol having the highest activity give it some sort of preference for further testing?
Does a dioxygenase not being able to use a certain substrate, limit its potential or ability? How is PhnA dioxygenase effected by not being able to use anthracene?
I found it unique too because it may suggest an entirely new group of genes that are similar yet distinct to the ones we already know about. I can imagine that alpha and beta subunits can often share similar phylogenetic trees having close to the same function in protein folding.
Tara I was wondering the same thing about the implications that this ability could mean for us. I know that arsenic is deadly to humans so us being able to use a bacteria’s/archaeon’s natural ability to utilize it could help our survival against it.
I was wondering the same thing Nolan. I thought it was noteworthy that the researchers even used the word “surprisingly” as if they also expected to obtain a PCR in the first place.
This may sound dumb but what is the purpose of the researchers going into full detail about the appearance and location of the hot springs they used in testing? Does it allow for future researchers to find as similar samples as possible or does it allow there to be no loose ends or grey areas in our current researcher’s methods?
Sarah Grace, I would also love to see a phylogenetic tree based on the arsenate respiratory reductase genes to potentially see the other bacteria and archaeon that’s able to use arsenic as an energy source.
Based on figure 1, it looks as if As(V) and As(III) are inverses of each other in the light to dark reduction of As(V) and the light to dark oxidation of As(III). This trend kind of makes sense to me in that for most redox reactions, there’s an equal reduction for every oxidation. The paragraph below suggests that both the reduction and oxidation of As(V) and As(III), respectively, are capable of cooccurrence.
The finding involving Ectothiorhodospira is good, right? Kulp being able to find an isolate of the genus that’s able to grow by photosynthetic oxidation of arsenite in an anoxic environment suggests that the 16S rRNA gene of the studied bacteria is similar to that of a known arsenite user. It was also interesting that the archaea genes were more diverse but they all seemed to originate from hypersaline environments (harsh conditions common of archaea).
Hey Sarah Grace, I thought it was cool too of how the rate of cycling was diminished. Your comparison of it to our own aging processes is very unique and comforting that we’re not the only organism to always be slowly dying lol.
What is the significance of the different suggestions of the arrA clones? Like the ones that compared closely to Ectothiorhodospira strain PHS-1 vs. the ones that didn’t indicate what biofilm microbes were associated with them?
Hey Tim, I was wondering also how effective transcriptomics or bioinformatics would be in a situation like this. I wasn’t sure where either could be applicable, but learning about these testing methods in lecture has made me wonder where they can be suited.
Hey Hang, that’s a super interesting concept to think of because I would think that activity could be present and similar results replicated in a different environment, following similar conditions. I would also think that controlling for those conditions in a new environment, especially one that is open and very dynamic as opposed to a lab setting, would be incredibly difficult.
No studies have investigated PAH-degredation from the region mention in the Arabian Gulf. Why is this? With studies in other areas, such as, the other sides of the Arabian gulf and the Kuwait and Iranian coasts, wouldn’t it be beneficial to add the data from this region to those studies and view/study it comprehensively?
I had the same thought Julia. I wonder if the initial extractable PAH value was a significant amount higher than post degradation.
Would light and scanning electron microscopes be the only valuable microscopy for this experiment.Would transmission microscopy be a advantageous tool for the experiment’s results or would it contribute little to the interpretation of data?
The article says that low concentrations of N and P are present and the environment has a high toxicity, which might limit productivity. How would it be definitively proven that is the N and P concentrations have a limiting effect and not the toxicity alone. Also, going off of that, how would one alter the concentrations and toxicity in order to reverse the inhibiting effect?
The last sentence says that OD is not suitable for larger size PAHs as a growth assessment method. If this is the case, why was implored? Is it because initially size of the PAHs were not known?
It is mentioned that O. intermedium, P. citronellolis, and Cupriavidus taiwanensis are less researched in regards to PAH degradation. I can’t help but wonder if there is a reason for this or if it has just been overlooked until now. Moving forward from this experiment, do you think that more researchers will take interest in these strains?
I agree with Danielle’s comment, PAH degradation seems to be under-researched. I have to wonder if this is due to financial limitations such as not enough funding or if it is a lack of interest in the topic, or something else. It seems as though this topic could be a major ecological boost for many ecosystems.
It seems to me the goal of this research group was to only identify and describe the different types of PAH degraders in the Guaymas Basin. Will research of this nature eventually lead to the cultivation of PAH degraders in areas where oil-pollution is high?
At the end of the paragraph, it is mentioned that the the relish spot are oil droplets. Is this color difference due to the high concentration of the oil in that specific spot, also is sites with that high concentration of oil targeted?
What is the purpose of the washing with fresh ONR7a?
Madelyn, I had the same thought. If this is true, shouldn’t the two sample have different traits in order to make it possible to be present at these deep depths? And if variations how present, does this mean the two samples are actually both cycloclasticus?
What is the disadvantage of carbon13 being distributed among other members of the microbial community? does it skew results or make it difficult to interpret results?
I agree completely with Jessica, as a skeptic, I always enjoy when results can be confirmed through one or more methods.
So just because they have differences at other loci, doesn’t mean they are different and this is due to their similar 16S rRNA genes?
With only 2 selenate reductase genes characterized, is this a growing area of research or is this topic often overlooked?
What is the importance of the discovery that the selenate reductase in E. cloacae being a membrane bound heterotrimeric comple?
Hi Melanie, I had a similar question. What causes the difference in color between red or white? Were the colonies grown on the plates similar to the ones we use in lab (blue colonies/white colonies) or is this color a characteristic of the actual bacteria?
What would be another method of verifying the FNR gene regulates Se(VI) reductase activity, other than a knockout mutation?
The first sentence in this paragraph states that it was hypothesized that the FNR transcription factor regulates selenate reductase activity. What might other possible regulation factors be?
The last sentence of this paragraph states that fermentative growth on glucose allows for reduction to occur while anaerobic growth on nitrate inhibits it. Would a possible expansion of research be to determine whether the nitrate itself is limiting the growth or whether the anaerobic growth is?
This first paragraph does a great job at identifying the problem and explains why this topic is important. When discussing oil spills I usually think about the effects on more large and notable animals (such as ducks or small fish). However, these environmental pollutants harm every aspect of aquatic life down to the micro biome
I think it’s really cool that scientist have possibly found a natural way to reduce pollutants in the soil. I understand that no foreign bacteria could be introduced into the soil seeing this could easily make problems much worse. I am curious on the possible outcomes of increasing bacteria that’s not very plentiful in the soil. What are the possible negative effects of this on the micro biome?
I’m not going to act lol a fully understand everything happening here. But i do find the process of PAH quantification to quite interesting. It so different from any quantification method I’ve ever used in a lab.
Here’s the authors do a good job of explaining why their research is important and how their particular research made a meaningful contribution
Here the authors do a great job at pointing out the possible sources of error. They noted their bias to the three strands of focus in their research and noted how that could have negatively effected the outcome of the experiment.
Although a lot more needs to done the results from the experiment seem to be very promising. I’m excited to see how this will be applied in a real world setting.
It’s interesting how some strains reacted to the diesel and others did
Do each of the common naphthalene-degrading bacteria noted degrade naphthalene to a similar percent in a similar incubation period?
Are there certain tests that are done to determine the best and most established strains for naphthalene degradation?
Was there a specific reason those 3 contaminated sites were chosen? Would it benefit to collect a sample form farther into the gulf rather than 2 from an island and 1 from a shoreline?
I know that hexadecane is an alkane hydrocarbon, but was there a specific reason this particular hydrocarbon was used for emulsification?
From 200-400 ppm, N1 had a greater growth than N10. I thought it was strange that at 500 ppm, the N1 and N10 strain were at equal growth, and at 600 ppm, the N10 strain had greater growth. No other strain experienced a change like this. Is there a specific factor that caused this change?
As stated in paragraph 9, N7 had BATH of 73.57% which was almost 7% higher than any other strain. Is this cell cur face hydrophobicity a key factor in why this strain had the highest level of growth even though N1 had the highest percentage of naphthalene degradation and E24%? Would the N7 strain be favored for bioremediation?
What exactly about the structure of a gram-negative bacteria makes it able to tolerate PAH uptake better than gram-positive which has the strengthening agent peptidoglycan? Is it because the fatty acids are bonded through the amine groups from glucosamine phosphate, or is it because of the different functions of the proteins in the peripplasm?
In the results section, I made the assumption that high cell surface hydrophobicity must be a stronger factor than emulsification activity because the N7 strain was more successful in growth than the N1 strain. I see now that there is a direct relationship and both are important in choosing the best naphthalene bacteria strains.
N7 belongs to Sphingomonas. Does the fact that this strain was identified and reported to use naphthalene as a soul carbon and energy source have anything to do with why the strain was so successful in growth?
I had the same question as Menesha, and also wondered if the binding abilities of teichoic acid embedded in the cell walls of gram-positive bacteria was a factor for its acceptability.
I had the same question as Menesha, and also wondered if the binding abilities of teichoic acid embedded in the cell walls of gram-positive bacteria was a factor for its acceptability.
Are specific PAHs amounts concentrated in similar areas? Such as a certain PAH amount is in areas with high forrest fires, and a level of concentration is in areas that are industrial dense?
Are specific PAH amounts concentrated in similar areas? For example, is there a certain/ similar concentration amount found in areas with high forrest fires, or a certain concentration amount found in industrial dense communities that are similar?
Does the organic compound that is utilized by the bacteria effect the degrading?
I had a similar question, and looked up the recipe to this media. Apparently it is easy to prepare and provides a broad base of nutrients. It was formulated for studying lysogeny in E.coli and has even been referred to as “Lysogeny Broth”.
Ampicillin is not a part of the LB recipe. I know that it is an antibiotic that can be used to treat bacterial infections, but why is this specific penicillin antibiotic used?
I did some research on ORF’s and found that their start codons are usually AUG. Is the fact that this ORF’s start codon is ATG significant?
I thought it was interesting that the activity was similar in the PhnC’s that were fused and unfused. Does fusing not have an affect on the activity usually?
I had a similar question. Would an experiment with the same variables yield the same results with PAH degradation on chromosomes rather than the plasma?
I do think that it would conserve resources to have one pair to partner with many different deoxygenates. However, keeping conservation out of mind, would the experiment benefit from making specific ferredoxins and reductase for each deoxygenate the cell would encode?
I remember reading that reductase and ferredoxin were 2 components that were normally needed for electron transfer to the terminal dioxygenase, however, could there have been other enzymes used for this electron carrier system?
I know that anthracene consists of 3 fused benzene rings. Is this why PhnA dioxygenase could not use it as a substrate?
Is the type of inorganic substance it employs significant? Are there other substances other than sulfide or H2?
I had the same question and looked up that the diel cycle is a 24hr period where the chemical and biological structure of a microbial can change as a consequence of changing light intensity. Changes can be detected using microsensors in pH, H2S and O2. Detecting the rate of these changes reveals zones of greatest microbial activity.
Anoxic groundwater is stated to have a dissolved oxygen concentration of less than 0.5 milligrams per liter.
RFLP is a technique that was one of the first techniques used for DNA analysis, however after looking into it, I noticed that it is not used widely anymore. Why was is used for this particular experiment and was there an alternative that could have been used that would have been more modern and accurate?
Because the results were not clear, are there separate/ alternative tests to determine if the oxidation activity was due to the aoxB-independent mechanism, to rule out that it was not due to the inefficiency of the primers?
Why was the sulfide condition not tested in the acetate?
I also agree with their approach. I also agree with the fact that they started off with As(III) in higher concentration to be more efficient. I assume that they experimenters knew it As(III) would oxidize in light and would therefore get results faster.
Responding to my previous comment, this makes me wonder if the results would have partially differed if they would have started off with As(V) at a higher concentration in the light phase.
Was there a certain amount of time that each population was exposed to the specific temperature? Does the amount of time that it is exposed have anything to do with the result of the temperature range experiment?
Using a non-oxygen acceptor allows chemolithotrophs to have greater diversity, while sacrificing energy production. Sometimes the use of an enzyme is needed for oxidation. Would the use of one change the results of this reaction?
[“Oil contamination can generate detrimental changes in soil properties, including modifications in maximum surface temperature, pH, and carbon and nitrogen levels (Aislabie et al., 2004).”””Altogether, this results in a significant decrease in species richness and evenness, and a large decline in soil biodiversity of contaminated soils (Saul et al., 2005; van Dorst et al., 2014, 2016).”] Mentioning species richness, the number of species within a community, and species evenness, the commonness of species within a community, it is also equally important to discuss species diversity, that of which combines species richness and species evenness. Biodiversity is more than just the counting of species, but it rather encompasses the genetic variability among organisms within a species, the variety of different species, and the variety of ecosystems on Earth.
[“This process can be achieved by promoting the growth of endogenous metabolizing bacteria in contaminated sites (biostimulation) or by directly seeding contaminated sites with pollutant-degrading bacteria (bioaugmentation). Since the Antarctic Treaty impedes the introduction of foreign organisms into the Antarctic continent, bioaugmentation can only be implemented by the use of native microbes.”] Why is it that only ‘native microbes’ are allowed into the Antarctic continent? What effects would the use of ‘foreign microbes’ have on Antarctic’s contaminated soils?
[Briefly, non-polar compounds were extracted from culture media using two volumes of hexane and vigorous mixing for 60s.]
What is the reasoning behind using ‘two volumes’ of hexane? Did this make the extractions more concentrated thus increasing the excitation-emission spectra of each sample?
[Cultures were grown with agitation at room temperature for 5 days, and phenanthrene metabolizing strains screened by change in medium color from clear to yellow due to the generation 2′-hydroxy muconic semialdehyde, a degradation product of phenanthrene (Treccani, 1965).
After reading the line above from the text, I became interested in the degradation product of phenanthrene, 2′-hydroxymuconate semialdehyde; that of which, I soon discovered is ‘formed from catechol by the enzyme catechol 2,3-dioxygenase during the degradation of benzoates’ & is ‘hydrolysed into formate and 2-oxopent-4-enoate by 2-hydroxymuconate-semialdehyde hydrolase.’
Since Chemotaxis seems to not play a part in PAH degradation, does that mean the bacteria treated the PAH like every other degradation process? If that’s the case, is there a particular reason it failed to differentiate PAH?
Since 16srRNA identified the isolates of Rhodococcus erythropolis, S. xenophagum, and Pseudomonas guineae, why then was only Pseudomonas guineae unable to process the diesel fuel as an energy source? Why is it that all three strains are psychrophylic yet 2/3 were able to utilize diesel, would they not all be able to utilize diesel in that case?
Dayana, I agree! “Increase in temperature increases the rate of oil degradation by bacteria” makes sense considering bacteria are more likely to thrive in warmer conditions. Warmer environments such as farms, industrial sites, landfills, and/or onsite sanitation systems are bacteria-rich and would likely be beneficial when using a microbial bioremediation approach.
Abraham, similar to you: I was also curious about ‘biosparging.’ After a quick google search I found: “Biosparging refers to air injection at pressures and flow rates sufficient to deliver supplemental oxygen, but less than those required to volatilize significant contamination. Evidence indicates properly designed sparging systems significantly enhance both biodegradation and volatilization.”
I was unsure as to what ‘siderophore aerobactin’ was, but after googling I now know that ‘siderophore aerobactin’ is a bacterial iron chelating agent found in E. coli & is commonly found iron-poor environments, such as the urinary tract.
Ryne, I also had this question.
When I think of Vibrio, I immediately think of the ‘flesh-eating bacteria.’
“Following incubation overnight at 30^C, V. fischeri ES114 colonies harboring transposon insertions were selected by growth at room temperature on LM agar plates containing erythromycin.”
Knowing that erythromycin is an antibiotic, is there a reason they chose this specific antibiotic as opposed to other antibiotics?
Ryne,
I asked the same question in a previous paragraph as ‘erythromycin’ was specifically chosen & I am curious as to what influenced their decision regarding the antibiotics used within the experiment.
“A strain of Vibrio nereis encodes 6 receptors, but it cannot produce any siderophores (Thode et al., 2018).” Why is it that a strain of Vibrio nereis cannot produce siderophores?
“Moreover, both vibrio species produce a variety of public goods including extracellular proteases, chitinases, and QS autoinducers, all of which can be monitored in real time.”
Because I was unsure exactly what a “chitinase” was, I google searched to discover that “chitinases” are enzymes that degrade chitin & contribute to the generation of carbon and nitrogen in the ecosystem. Chitin is a component of the cell walls of fungi and exoskeletal elements of some animals (including mollusks and arthropods).
“It connects the recycling of organic nitrogen to the ultimate nitrogen loss from the oceans, because its products are substrates for denitrification and anaerobic ammonium oxidation (anammox), the only two presently known nitrogen loss processes.”
Anammox (anaerobic ammonium oxidation) is a reaction that oxidizes ammonium to denitrogen gas using nitrite as the electron acceptor under anoxic conditions. Anammox was an important discovery in the nitrogen cycle.
“It is responsible for the formation of the large deep-sea nitrate reservoir.”
Are hydrothermal vents the only cause of the surplus of nitrate in deep ocean water?
“A 20- to 40-m-thick suboxic transitional zone, characterized by low oxygen and undetectable sulfide…”
Due to the low oxygen and undetectable sulfide levels in the Black Sea, does life still exist within it? Are these levels toxic?
“Abundance of total Archaea was taken as the sum of cren- and euryarchaea.”
In efforts of focusing on the data collected rather than ‘how’ they collected it, would a high point within this paragraph be the equation: “[cren-] + [euryarchaea] = total Archaea abundance”?
In this paragraph, I feel as if this particular sentence holds importance: “15NO2 − production was measured in the same anoxic 15N incubation vials as in the anammox rate measurements, but 15NO2 − was analyzed as N2 after a two-step reduction by acidified sodium iodide and then by copper at 650°C.”
From the Figure 1 graph(s), I was able to gather that as ammonium levels increase, nitrate levels decrease. Additionally, the high anammox bacterial levels are indicative of nitrogen loss during oxidation.
From the Figure 3 graph(s), I was able to discover a correlation between the species of bacterial and crenarchaeal amoA expressed & the depth of the ocean; the species expressed falls as the depth rises.
It is really interesting that Diesel Oil is most commonly used fuel in Antartica when Antartica is considered to be one of the most untouched places on Earth. So to read that we are using one of the most toxic fuels there is disheartening. But I am glad that they are trying to help combat using other toxic things.
I am interested to see what their outcomes were especially because the antartic ecosystem is particular different than a lot of other common ecosystems in the world. Also the latest source from this paper was 2017. I would be interested to see what it is now in 2021. Have we made process with this? Or are we still harming our environment?
WOW! It is incredible that one could access the DNA sequence code online, and the fact that you could find something similar is amazing too. I also find it interesting that it took so much to get this approved/ submitted. I wonder if the process is this intense now? Especially because what they used was from 2000.
I wonder why after two months they were analyzed? Is that so they were for sure isolated and not contaminated? Does that happen a lot in other experiments? I understand it is coming from Antartica, but that is still an extremely long time.
It is really interesting that they are psychrotolerant rather than psychrophylic, when you think they would do better because of their hostilely cold environment. But it makes sense that it was seen previously because bacteria can get trapped in ice.
It is really interesting that it has to be under the right conditions to grow. One could see that they had a huge struggle with getting D43FB to grow in paragraph 8. But I am intrigued at how picky these things are by deciding what special environment they want to grow in.
I totally agree with you! I never would have guessed how many specific bacterias there are in soil. But also just in this experiment alone where they had what seems like an endless opportunities to find almost any bacteria. It is really hard to wrap my brain around how many bacteria there truly is.
I wonder if this is true with extreme environments in the ocean/space, or if this is just by land? Why do they have to be extreme environments, and what is the benefit? Also it makes one wonder that yes, what they are researching is amazing but is there an easier/ cost efficient way to travel to these environment and take samples?
This is incredible that bacteria is abundant. This is insane that these are responsible for recycling of the hydrocarbons. Especially because these things are not even on the surface.
How many other bacterias are there and how much does this cost? Also, how does this help us in the future because finding more oil-rich sediment is great but how long until this taken to use for resources? I think I understood this correctly, if not, can someone help me understand better?
does pressure play a part in this? Does it matter how they keep it in lab? No pressure has to be applied to the tubes? Even though this bacteria is found at extreme depths?
I am interested to see which temperature works because 4 degree C and 21 degree C are two completely different temperatures. So I’m curious as to why so high and so low temperatures to test. Is there no in between?
I wonder if the amount of oxygen had something to do with why they could not cultivate. Bacteria at the bottom of the floor verse in the shadows uptake and use different oxygens. So, maybe the cultivating has something to do with how much oxygen they get.
Referring to the last paragraph as well, I am curious to see if the shallower bacteria degrade oil like the deep sea because I know oil floats but I also see oil in shore too as well from spills and what not. So I would like to research more on that too.
Interesting that the process of nitrification and denitrification affect it so much. Especially because we do not hear denitrification getting used that often.
Just to note the archean Earth had methane droplets, there was no oxygen. Oxygen was only in water. Just a reminder of what this time was like.
I have actually drove past this lake when I was in California, I had no idea about the springs and what not.This is so crazy, it was crazy in person too!
I am wondering why they failed to show significant similarities when it seems like they would. I wonder what would happen if the testing did go on further?
It is really interesting that it had the opposite effect. I wish they would have shown the data for the rest of it.
I wonder if it would have happened if they had the primers for this particular application.
Wow this is so amazing! That even to this very day and in present experiments this pattern still happens.
I wonder what the novel mechanism is? I need to do more research on how As(III) microbes can provide more knowledge for us and what that means?
On the graph you can see that one is clearly much higher than the other others. I’m wondering what went wrong with the others?
They talk about using the heavier DNA, but then acknowledge the fact that the lighter DNA could have the reaction to degrade it too? A lot of these ending results experiments seem to pick the better one but then note that the other data not used could be it. Who is studying this other weaker experiment? Will it get studied?
Nothing was mentioned that said that these naphthalene-degrading bacteria were introduced to the Persian Gulf. Does that mean that these species are native and the purpose of this study is to identify those that thrive in these PAH- and oil-rich environments to further study them, or is the purpose to identify which species that they introduced was performing best and in which environments?
Is the PAH bioremediation really environmentally benign? Granted, it fixes the pollution problem, but doesn’t it create a dead zone (which is an oxygen-deficient area) from the formation of CO2 and H2O (from both the breakdown of the PAHs and presumably the use of O2). Also, the creation of the microbial mass would be similar to algae blooms in that when those microbes die off, it creates a large food source for the decomposer, which use O2 to feed on that microbial mass, thus also creating a dead zone.
What is emulsification activity, and what do the results tell us about naphthalene-degrading bacteria that they are isolating?
What is the purpose of doing the estimation of the remaining naphthalene when a more accurate GC measurement was taken later in this methods section (paragraph 11)? What does this version of the measurement tell you that the other does not?
It was mentioned earlier that the significance of emulsification activity and BATH test determine how well a bacteria can attach to a carbon source. I can understand the surface hydrophobicity (because the bacteria’s hydrobicity can be used to attach to the PAHs hydrophobic region), how does emulsification affect the bacteria’s ability to attach to the PAH source?
So when these scientists discover these strands, even though they are showing the general genus and species that they are related to, is this the equivalent of someone discovering a new subspecies of a known animal? Also, how different can different strands of the same species be?
If they are trying to determine a bacteria’s ability to break down PAHs, why are they using naphthalene as the determining food source? Shouldn’t they use more complex molecules with more benzene rings to make it more selective since PAH stands for polycyclic aromatic compounds, which indicates that any multitude of benzene rings could be present in a single compound. If they just base their research off of naphthalene doesn’t that mean that the more complex segments of the PAHs could be left over?
The Gram-negative is what I would have hypothesized to do best because it has that outer-membrane. Why, then, did the last experiment conclude Gram-positive were the naphthalene-degraders? Were there no G- present in that environment, or did they just not break down naphthalene? Also, did the G+ degraders have special adaptations that allowed it to tolerate these types of environments without the outer membrane? What could some of those adaptations be?
I thought there was not much difference in the ability to break down a simpler PAH such as naphthalene compared to the more complex PAHs that they are talking about, with the only difference being that it takes longer. If that’s the case, then why are they distinguishing between naphthalene-degraders and other, larger and more complex PAH-degraders?
How are these genomic sequences able to determine whether this strain is a good candidate for performing bioremediation? Are these sequences compared to known sequences of organisms that are capable of degrading, or do they say something about the speed and/or ability of this strain to perform the necessary catabolic pathways to break down the complex PAHs?
Is there a way to separate out the enzymes to determine what each one does specifically in order to identify the ones involved in the PAH break-down? Or do we have to just look at the activity results from a solution containing all of the enzymes and just be able to say that at least a few enzymes are present in the cell that are proven to break-down PAHs?
Why are they looking at the plasmid DNA sequence? If the genes encoding for PAH-degrading enzymes are found in the plasmid, does that mean that this bacteria doesn’t naturally degrade PAHs, but rather got these sequences from another organism present in the soil (and maybe even from on of the other organisms from the original soil sample)?
They are testing for all of these different aromatic compounds, but how many different types of PAHs can actually be found in oils, plastics, or other potential PAH-contaminants? And, just out of curiosity, on average how many aromatic rings do they have?
So is the purpose of determining if two ORFs (such as phnH and phnI) are detected next to each other to see if they could encode for proteins that are part of the same biosynthetic pathway? Or is it for another reason?
What does it mean by CDNB-accepting? Caroline and Marissa said that GST was use for detoxification, so does that mean CDNB is a toxic component of phenanthrene, or is it a toxic by-product of the breakdown of phenanthrene, or does it indicate something else?
Is there a reason that they needed to compare both sequences? Was it because the 16SrDNA sequencing isn’t accurate enough, or did they just want to make sure they classified the strain correctly?
Why are they focusing on meta-pathways for degrading PAHs? Is there an advantage to having that sort of pathway compared to another type like ortho-cleavage pathways?
Just a thought, but, does the ability of bacteria and other unicellular organisms to undergo horizontal gene transfer affect the accuracy of determining phylogeny based on genome sequences, since they can transfer some of their genome to others not necessarily of the same species or genus?
Is it more common to find As(V) reduction and As(III) oxidation coupled between different species or coupled within one organism (as in one organism can do both)? Also, is this paper going to focus on the relationship between an As(III) oxidizer and an As(V) reducer or on a single organism that can do both?
I know hot springs (which is created through volcanic activity) also contain H2S in addition to the As(III), and I know there are many microbes that use H2S in the process of chemosynthesis. Since these microbes use the As(III), does that mean that there is a selective advantage to using As(III) for photosynthesis rather than H2S in chemosynthesis?
Where did the Ectothiorhodospira strain PHS-1 come from? Is it a known As(III)-reducing microbe that is acting as a control?
Why were the biofilm cells washed before being added to the anoxic medium? What does that remove?
What was the point of comparing the Ectothiorhodospira strain to the rest of the data?
Why would the assimilation of acetate not be stimulated by the addition of arsenic oxyanions in light conditions if it is the reduction of As(V) that helps drive the oxidation of acetate? Shouldn’t more acetate be assimilated if more As(V) is added?
Can you really say that the PHS-1 strain only does the oxidation of As(III) to As(V) if the PHS-strain is only measured for the As(III) rate rather than the the As(V) reduction rate (since the graph only indicates that under the light reaction that this oxidation takes place and does not consider the As(V) reduction)? Shouldn’t this strain be cultivated in the dark to truly determine that the reduction if As(V) does not take place in this strain?
How do they know that the rates of the oxidation of As(III) and reduction of As(V) decreases over time? Is there another study that they are referring to?
How did they not know which microbe was able to perform the As(V) reduction if they were able to isolate each Bacteria or Archaea strain (or did they really isolate each strain?) and test them on their performance in a dark environment? Couldn’t they just measure the amount of As(V) that was reduced in the dark environment to determine the rate of reduction?
I find it interesting that we are exposed to PAHs through ingestion; however, substances such as glucose, yeast extract and broth aid in the degradation of PAHs. What foods contain PAHs? Is it public knowledge that these foods are possibly harmful?
I agree Madeline. I am very excited to see our results as we isolate and characterize naphthalene degrading bacteria from local soil samples in the lab!
I enjoyed using centrifugation and chromatography in prior chemistry labs. Will we be using these techniques in our lab as well?
I find it interesting that this study used a soil sample from an abandoned oil field in Japan, while we used a soil sample from the base of a light pole in Dr. Ni Chadhain’s neighborhood. I guess this just shows the abundance of PAHs in the environment.
I was curious about this initially, as well. However, after reading the entire article, I realized that the added yeast extract in YMSM plates increases the rate of PAH degradation.
I was also curious as to why the samples were continually being shaken at 180 rpm for two weeks. I did not know that. Thanks for sharing!
I wonder why Fla was degraded significantly more than Nap and Phe in both the MSM and YMSM?
What caused the 24 hour delayed degradation in MSM as opposed to YMSM?
I was curious about the clear zone formation technique. I looked it up and found that the clear zones around the colonies represent its ability to inhibit other microbial growth.
What caused the complete inhibition of Nap degradation by the addition of YE in the medium?
I recall the last paper stating that PAHs can be introduced to humans via dermal uptake, respiratory uptake or ingestion.
If obligate marine bacteria are possibly more significant PAH degraders in costal marine environment rather than terrestrial bacteria, why is there only a limited amount of research? It seems that this could be substantial to the health of marine life and, therefore, human that consume seafood.
I never really considered purchasing the chemical and reagents. I guess I just assumed that the researchers obtained and/or isolated them independently. So I found this interesting!
Due to its high conversion rate, is 1-Methylnaphthalene a more sufficient substrate than the others listed?
I find it fascinating that we are able to clone specific genes in DNA and use them to our advantage! I wonder if this could be a permanent solution for the PAH crisis?
I wonder why the relative activity level of PhnC with fused, unfused and monocyclic catechols were nearly all the same?
Louie, I’m a little confused by this as well. I reviewed my notes from class yesterday and noticed that I wrote “primer walk” following the direction of the arrows down the nucleotide sequence.
I wonder why PhnA was not able to convert anthracene and monocyclic aromatic hydrocarbons, but is otherwise able to degrade a wide range of aromatic hydrocarbons.
I find it interesting that the 3D structure of the oxygenate component may provide access for substrates to catalytic iron on NdoB.
Spencer, I too was curious about the function(s) of ISP. So I did some research and found that they fulfill functions in electron transport, enzyme catalysis, homeostatic regulation and sulfur activation.
I was also curious about this and was wondering if this ability could be the result of a previous genetic mutation?
It is interesting that this paper, in contrast to the first two, studies the characteristics of both bacteria and archaea. I am curious to see how the remainder of the paper will be presented.
Hi Derria. I was a little confused by this at first too, but I looked it up and it seems to mean a 24-hour period. Here is the definition I found: “of or relating to a 24-hour period, especially a regular daily cycle, as of the physiology or behavior of an organism.”
Jesse, I was curious about that as well. I think that the sequences that did not show similarity were excluded because they would not be useful in the remainder of the experiment.
I was also wondering if the times of the sequencing were significant. The gap between the first two months was only two months, but the gap between the last two months was seven months.
Alex, I too find this interesting. Like I mentioned earlier, I wonder if time could be a factor? And if so, by how much would the results change?
I find that interesting as well, especially since we’ve primarily studied bacteria thus far. I also suspect that the harsh environment contributes to the larger number of archaea vs bacteria.
I find it interesting that the genera of the nearest neighbors of representative clone are so diverse, because the top 10 matches on both of the BLAST searches I performed in lab were all from the same genus.
Is it common to observe oxidation of certain microbes in the light, but not in the dark?
Dawson, I also found this interesting. Microbes’ ability to just change their properties as needed is so fascinating to me.
Unfortunately, with any highly visited area there is always going to be pollution affecting the natural habitat of the area. It seems like these explorers do not have any consideration for the environment that they want to visit. Maybe a solution would be to restrict the amount of people that come to Antarctica per year and put rules in place to designate what kind of equipment that can be brought and where transport of vehicles is allowed to minimize the areas of oil spillage.
It is very amazing to see how scientists can use the bacteria from the Antarctic soil to degrade a key component of diesel-fuel. This allows to them to help fix some of the damage that has been done from the oil spillage. I wonder if these scientists have looked into using electric vehicles as an alternative to transport and if the effects are less detrimental. As long as these diesel-fuel trucks are coming through at high volume, it will be hard to tackle to effects of oil spillage.
This makes me wonder what environmental bacteria was collected that is unique to Antarctica. Due to the extreme weather conditions, I imagine that the microbes collected would be very different compared to the ones collected in the climate experienced in the United States. This also poses the question that if the environmental bacteria in the freezing temperatures of Antarctica are unique, are they more susceptible to the negative affects of diesel fuel?
The crystal violet solution and the use of an alcohol sounds like they could be doing a gram stain on the bacterial isolates. However, in this case, crystal violet was used to measure absorbance and assign the correct density of each culture. Is this another way of saying they used a gram stain to determine the thickness of the peptidoglycan layer, or is this another analyses altogether that just happens to also use crystal violet and an alcohol?
It’s interesting to read how scientists are able to observe if chemotaxis occurred. Instead of having to view motility under a microscope, it seems that they were able to determine the ability of chemotaxis by seeing if the bacteria were present closer to a target compound, where they previously were not. This paragraph gave me more insight on how lab processes are done.
The finding that the presence of heavy metals reduces the degradation of diesel fuels raises more questions about the environmental pollution. Is it possible that the traffic going through Antarctica leaves traces of metal from the vehicles and equipment along with leaving traces of diesel fuel? And if so, will the use of these bacteria be impactful on the diesel fuel or, instead, become a failed attempt?
This paper taught me that scientists are able to modify bacteria to enhance their capabilities. It seems extremely advantageous that this is something possible to further degrade the diesel fuel and ,in turn, help the environment. Could formation of biofilms, secretion of biosurfactants, or the ability to form chemotaxis be enhanced to enable the bacteria to work more effectively with the presence of heavy metals?
This paragraph is excellent at explaining the way that these scientists cultured the bacteria and how it can be further researched. Hopefully this paper inspires others to experiment with different culture techniques and will enable them to possibly find more types of PAH degrading bacteria. The more research that can be done on the different bacteria found in Antarctica, the more of an increased likelihood that the diesel fuel pollution problem can be overcome.
It’s interesting that scientists are able to spray on the growth substrate which then leaves a layer on the agar for the bacteria to use. This seems like a quick and efficient way to ensure the bacteria have their source of carbon that is evenly distributed.
It’s interesting that scientists are able to spray on the growth substrate which then leaves a layer on the agar for the bacteria to use. This seems like a quick and efficient way to ensure the bacteria have their source of carbon that is evenly distributed.
Was there oil-degrading bacteria found in these samples? Or were these oil droplets taken just to study the oil properties on the seafloor?
I wonder if 11 days in lab time is considered long or short to wait for sufficient results. Also, is there some type of enzyme that can be added to 13C to make the degradation and mineralization process faster and more efficient?
Now that the scientists have found that Cycloclasticus are obligate PAH degraders, it would be interesting to see how a scientist would use this information to enhance the environmental conditions of the sea floor. Are there bacteria similar to Cycloclasticus in other parts of the sea that are affected by oil or would these areas benefit from having scientists plant the founded PAH degraders in parts of the sea floor other than Guaymas Basin?
Looking at the figure, I’m assuming that all of the PAH degrading bacterial strains isolated are from the same ancestor. I wonder if the related strains from GenBank that are given would have the same or better PAH degrading properties? This might be good supplemental research to pursue.
Is the rusty color produced from some type of by-product from the chemical reaction taking place? Also, is this by-product harmful to the environment but just less harmful than oil that is not degraded?
When recreating the hot spring water for incubation, it says that O2 free N2 was dispensed in anoxic tubes. So, it’s safe to assume that the hot springs created an oxygen-free environment for these bacteria and archaea to live in? Or is this just for the purpose of the experiment?
I looked up crimp sealing and couldnt find an exact definition. I’m guessing it is some specialized seal on the serum bottles to keep the gases and samples inside and any microbes or dirt etc outside to keep the samples just as they were when collected. It also might be important to use to keep oxygen from interfering with these anaerobic microbes.
Shelby, I was wondering the same thing. I would also assume light could withstand a higher temperature. Maybe its possible that they needed a more specific temperature with light to be able to use it as an energy source.
It’s interesting that both As(III) and As(V) have almost the same optimal temperature and general shape of curve. The difference is that the dark incubated biofilm has a larger range of temperature to be able to oxidize as well as a slightly higher oxidation rate for each point than the light-incubated counterpart. Due to the close similarities, I wonder if the oxidation is actually the same for each mode of incubation, but possibly there are other aspects of the experiment that allowed for the dark-incubation to have a slightly higher oxidation rate.
I wonder if at 50 degrees C and over these microbial populations start to die off. Maybe the temperature that the biofilm is active is the window for which the biofilm can survive.
I wonder at what point did evidence of oxygenic photosynthesis arrive? What microbes that utilize oxygenic are the closest in relation to these arsenite degrading archea and what mechanisms do they endure that are similar?
I like how the introduction of this paper talks about how oil-degrading bacteria has been explored much more recently and that the sea floor would benefit from this type of research considering the oil spill in the Gulf of Mexico. I think we rarely think about the long term effects from the oil spill and that there’s still clean up to be done. I’m glad that research on oil degradation is being done in this regard.
It’s interesting how scientists are able to studying these hydrocarbon-degrading microbes with the use of SIP. I wonder where else these specific bacteria live other than off the coast of California and if it is possible to place these bacteria in different areas of the sea that they are not originally found to degrade oil.
The prior papers studied phenanthrene degraders as it relates to oil pollution and could be environmentally useful to understand the bacteria. For this paper, I understand that they are studying how the bacteria and archaea utilize concentrations of the different forms of arsenic, but what was their driving factor? Was it just for the sake of understanding it better or to use this research as a way to degrade arsenic when needed?
In the photosynthesis dealing with water and sunlight that we normally learn about, water is the electron donor and light is the form of energy. Anoxygenic photosynthesis is photosynthesis without water. So, these specific bacteria are using As(III) as their electron donor in place of water?
From this paragraph, it seems that the researchers received the opposite result that they were expecting regarding the samples’ ability to degrade hydrocarbons. They explained this, however, by attributing sample 4571-2’s lack of hydrocarbon-degrading ability to its use of sulfide instead of oxygen, an adaptation to anaerobic environments.
By using the picture and graph, we can interpret that there is one gene that is more abundant than the rest. That gene was in fraction 6-10, which means it represents 13^C heavy DNA, indicating that 13^C heavy DNA is found in high quantities.
After reading this paragraph, I was reminded of a personal assumption I made that in nature, bacteria lived closely to bacteria that have similar genetic makeups (offspring stay close to parents). It is interesting to me that bacteria that differ so much genetically, observed in their ability to degrade PAH and possibly morphologies, live in such close proximity.
I am interested in the reasoning behind the -20 degrees Celsius temperature i which the agar slants containing isolated strains were kept in. Was this to prevent colony growth and reproduction, as bacteria grow and reproduce more quickly at a warmer temperatute?
What did the phylogenetic tree tell the researchers about the axenic isolates? Did they’re evolutionary relationship to certain bacteria suggest a reason as to why it is the result of pyrene being used as a carbon source?
I am interested in whether the Gram test was positive or negative. I would also if the physio-chemical properties of the isolate’s cell walls play a role in determining what PAH the bacterium use as a carbon source.
Does BC1’s relation to O. intermedium suggest that it, too, effectively degrades high molecular weight PAH such as pyrene and benzo(a)pyrene? BC1’s relation to Brucella melintensis also suggests that it may be present in livestock and humans; could this information be applied to health or agriculture studies?
Isolates LB and LC are clearly different in that LC is a hydrocarbon degrader and did not grow in the PAH while LB degrades citronellol and grew wonderfully in the PAH. What about being a hydrocarbon degrader made the LC so incompatible with the PAH environment, or what about being a citronellol degrader made the LB so compatible with the PAH?
The basis of petrochemicals are hydrocarbons, which explains why this environment for ideal for studying hydrocarbon-degrading bacteria. However, once those petrochemicals are broken down, what are the recycled into?
It appears to me that the SIP technique has been utilized for identifying microorganisms and assigning them to phylogenies based on their ability to absorb a stable isotope. This does seem like it would be very useful if one of the goals of this experiment is to identify the number and diversity of PAH-degrading species there are in the samples.
As shown in the captions of the pictures, one was taken at the sampling site for background cores while the other was at the site for hydrothermal cores. What is the difference between these two?
I recognize SIP as a technique used in the last article to distinguish bacteria based on their ability to degrade hydrocarbons. It seems that certain sequences of DNA in these samples were identified by SIP, suggesting that these samples help the bacteria in biodegradation.
It is very intriguing how the enzymes in the different bacteria, which are made based off genetic code, could affect the SE(0) product structure. I think I understand this correctly, but the reduction mentioned in the first sentence is referring to the use of selenium oxyanions as terminal electron acceptors in anaerobic respiration right?
As there are two mutagenesis methods we have discussed, I wonder which these researchers picked from. I believe the best one to use would be site-directed because on of the objectives of the experiment is to identify important genes.
The LB medium that the bacteria was originally grown in contains many nutrients that promote bacterial growth, while the minimal salt medium would only allow the growth of bacteria that could use the included compounds as an energy resource. Transferring the bacteria to this selective medium was intended to isolate the target bacteria, correct?
If I am reading this correctly, the wild type of the cloned DNA was Km and Gm resistant, so when plated on LB agar supplemented with Km and LB agar with Km&Gm, growth on the first but lack of growth of the second indicted a mutation. Would that be a mutation in the Km-resistant gene?
I believe that is correct, that the minimal salt medium would separate the types of bacterial colonies by only allowing the certain bacteria to grow. Those that grow then produce red as a result of the selenium reduction.
A knockout mutation is one in which a gene is made inactive. This shows that any protein or function exhibited with that gene and then consequently not shown without the gene could be controlled by that gene; however, these researchers have to address that another reason might explain the lack of exhibition.
Since the regulatory genes from other Sphingomonas strains that degrade different PAHs have been isolated, would it be possible that these regulatory genes could be similar enough to give some insight to finding those associated with acenaphthene degradation?
From my understanding, it seems the gene cluster that was isolated is located on the same segment as the arhA genes, but these are not all of the genes used in the process of degradation of acenaphthene. The rest of the genes are most likely scattered.
Does the fact that the Gm cassette and arhR and ORF15 are transcribed in the same direction help the plasmids bind to the DNA for gene disruption?
Why do they choose to amplify the regions between arhA3 and ORF6? Are they looking for a change in expression levels of the region between?
From my understanding of previous papers and discussions, the initial oxygenase enzyme is the one responsible for the production of indigo from indole. So, if they have lost the ability to produce indigo, it could be due to the absence of the gene/gene product.
What is the benefit of using a spontaneous mutant to insert the plasmids into versus an E. coli host?
From my understanding, I think they are highlighting the structural similarity of fluoranthene to the other PAHs as being the reason that their biodegradation pathways are similar, not necessarily the same.
Celeribacter indicus P73T is the first fluoranthene degrading bacterium found in the family Rhodobacteraceae. They are studying this bacterium because of this and I think they are using fluoranthene because it is a good model PAH when studying PAH metabolism.
I understand sequencing the entire genome is a costly process, so what was the main benefit to doing this first?
Does analyzing the intermediate metabolites of degradation of fluoranthene give the researchers information towards which kinds of pathways are being used, depending on the types of intermediates?
Were the PAH degradation genes acquired by strain P73 via horizontal gene transfer located on a plasmid or integrated into the chromosome?
From my understanding, the increased percentage of genomic DNA encoding transporter genes in strain P73 is a result of the strains ability to degrade PAHs.
Is it possible that strain P73 lost the genes encoding Fli and FliS, and subsequently lost its flagella?
What is the advantage of strain P73 having a gene for sensing aromatic compounds?
So, they were able to determine the pathway through a combination of computational predictions and metabolite analysis? Other than the metabolite analysis and the gene deletion, their entire approach has been computational predictions, correct?
Would the research approach they used here be considered a preliminary attempt to answer the kinds of questions the last few papers have answered? For instance, this group analyzed the genome and proposed a possible metabolic pathway, but did not perform any further testing to prove the proposed pathway.
Before reading this paper, I wasn’t very informed on this topic. I have learned a lot about the toxicity of PAHs. The statement I found most interesting was that the catabolism of aromatic compounds in rhodococci leads to the transformation of these compounds (found in PAHs) to TCA intermediates. I wonder if we could somehow manipulate this ability and think of PAHs in a positive way?
Based off the comment, “Fla…has potentially carcinogenic effects,” I am curious to know how much of a role it has in cancers studied today? The word “potentially” also stands out to me because I wonder if it hasn’t been studied enough to draw legitimate conclusions or if this statement is only relevant under certain conditions like temperature, part of the body etc?
Hi Jesse! Yes I think that it is safe to assume that Rhodococcus sp. is the better option with regards to this experiment because it neglects to mention otherwise. It also states, “the capability of Rhodococcus sp. to degrade high concentration of Fla does not exhibit homology with catalytic domain of previously reported ARHDs.” I interpreted that statement to mean that it was a better option!
I’m not entirely sure, but I think that it is a way to “purify” a sample of bacteria. Maybe it could be a way to control for unwanted growth during the experiment?
I’m not entirely sure, but I think this could be a way to “purify” the bacteria. Maybe the bacteria is washed to control for unwanted growth during the experiment?
I wonder how running the PCR product on agarose gel helps us to confirm apt purification? As someone who hasn’t run a PCR before, I wonder if that a “normal” way to conduct an experiment similar to this one or if this gel was chosen for a different reason by the experimenters.
I think this part of the results section is one of the most interesting. It’s amazing that we can use DNA to try and identify the different bacteria strains in a media. I think those percentages are pretty high and demonstrate good accuracy in the identifications.
The extent of differences between these three PAHs was interesting to me. I assumed Fla would be degraded more than Nap and Phe (which happened) but the individual percentages and the increase in residual PAHs for Nap YMSM surprised me.
I agree Dawson! I am also interested to know how popular rhodococci is as an organism in biology studies today. In other words, is this is a common organism molecular microbiologists use in research? Is the sentence, “rhodococcus is one of the most promising groups of organisms” a “given” in molecular microbiology research?
I agree Justin! I also find the sentence “there are studiess reported where bacterial strains degraded HMW PAH but not LMW PAH to be very interesting. I would have assumed the bacterial strains would be able to degrade lower molecular weights to an easier extent. I wonder what caused this to happen? Was there a sort of affinity to the HMW PAH that was lacking in the LMW PAH?
I’m curious to know how the different solvent systems in the thin-layer chromatography column effected the purification of the converted products.
I think that this part of the experiment was really cool especially because we have discussed the lac promoter in class. Also, further research in our ability to manipulate genes in order to become over-expressed (or repressed) could have really interesting effects on various genetic diseases.
From what I understand, the ability to oxidize indole to indigo means that there’s a presence of an aromatic oxygenase gene. Kind of like in lab when we saw cells turn pink when placed on the Macconkey agar. The color pink meant that they could ferment lactose. So, I think that that means our degrades have that aromatic oxygenase gene as well. I interpreted that statement to be causal.
I agree! I think that this paper describes really cool techniques that give us the opportunity to learn more about metabolism and microorganisms in general. I also wonder what the not continuously clustered genes mean in terms of degradation potential of PAH, especially with the mentioned high degrees of similarity with the sequences used for catabolism. In transcription with regulatory genes order matters, so I assume it’s the same concept for this.
I agree! I also find it interesting that both genes function similarly despite the different order of the phn genes. Based off what we have discussed in class so far, I would have assumed that the order of the genes would affect the overall function.
I think so! But I also wonder if having a shared dioxygenase system is a greater contributing factor to PAH degradation than genetics?
I find it very interesting that this paper resulted in the possibility that PhnC is involved in both the upper and lower pathways for degradation of nap, phe, and biphenyl. This seems like a very different result and approach to researching PAH degraders than our first paper.
I think that the topic of this paper is really cool because not only are there bacteria that can survive in the presence of high concentrations of arsenic (a known toxin), they also the ability to use it to help increase energy. I think it shows the truly diverse and “smart” nature of microbiology.
I think the focus of the examination of the cycling of arsenic under anoxic conditions is very interesting and important. Due to the Earth first being anoxic, this will truly help us get one step closer to understanding how organisms survived through the use of cycling of arsenic in that time period.
I agree; that sentence stood out to me the most. I would be interested to see how the researchers responded to the lack of the PCR product and how it impacted what they chose to do next.
That surprised me as well. In classes we never mention bacteria possessing that same characteristic of archaea. I’m interested to see if there are any differences in their respective ability to live in environments rich in arsenic.
I am curious as to why singlet clones are not generally used for sequence analysis. Is there a lack of their ability to be sequenced because their RFLP groups are represented by only a single clone?
I am interested to see what the radioassays of the chemoheterotrophic or chemoautotrophic processes resulted in. I am curious to see the difference between the two groups of organisms with relation to arsenic biotransformations.
I am curious to know if the experimenters knew that the thioarsenic intermediates were soluble at high pHs before the they saw the errors associated with the sulfide-amended samples. I am assuming they didn’t but maybe they did and the use of high pHs was the only way to get the samples?
While the observation discussed in this paragraph is important because it shows which organisms are the dominant arsenic cycling ones, is the detection of only one clone type a substantial find? Are there usually more clone types when conducting experiments such as this one?
I found this part of the paper to be very interesting. It’s really cool that accumulation of As(V) allowed a new set of niches to open. I also find it interesting that the cyclical reactions are able to show a shift between aerobic and anaerobic conditions even though it occurred such a long time ago.
I would assume that the next step would be to try a different set of primers because that seems like an “easier” solution than trying to isolate new types of aerobic As (III)-oxidizing microbes and then doing follow-up biochemical investigations. However, I think if the oxidation was being carried out by a novel mechanism, that the new information obtained could be interesting and provide scientists with important information that would help us understand the way systems work more fully.
This paragraph specifically mentions petroleum PAHs. Were they chosen to be studied for a reason or does the bacteria have stronger and more diverse degradation effects on petroleum PAHs compared to others?
I agree Lane! I also wonder how this investigation could effect the marine-located PAHs that humans have contributed to. This experiment could open up lots of research and discussion about how to potentially minimize the damage that we have done through our use and disposal of petroleum products.
I agree Anna! I think that their environments have helped them become more equipped to degrade marine PAHs than terrestrial PAHs. I wonder if the difference in marine and terrestrial physical and chemical environments (such as internal temperature) could have something to do with optimal PAH degradation activity as well.
Tara,
I had similar thoughts on somehow isolating the specific ARHDs and then researching their functions as it relates to the catalysis of PAHs specifically. Perhaps having the ability to garner this material would prove to be a more efficient manner of degradation.
I suppose that also brings about some other concerns such as financial implications, as you mentioned, and also the potentiality for negative impacts on surrounding ecosystems.
I, too, found it to be interesting that the addition of particular organic and inorganic supplements could potentially amplify the impacts of PAH-degrading microorganisms.
I’m curious to know whether or not some of those supplements are more or less effective than others, and if so, what is the reasoning behind that? Also, perhaps there is a particular supplement ratio (a certain amount of nitrogen and phosphorus) that would create a highly conducive degradative environment. Is it possible that in the presence of certain amounts of organic or inorganic materials there would be a negative result, as well?
I’m curious to know the reasons why Rhodococcus is more versatile in its ability to metabolize a broader range of compounds.
Is there a particular structural difference between this strain and the others mentioned prior, such as Mycobacterium, that render it more effective on this front?
I am curious of the significance of the development of chromarods in hexane and hexane/toluene. In what way do these two materials together influence this development? Does this mixture somehow help further isolate the desired material similar to the chemical used as an anti-fungal in lab 2?
What significance does nitrogen have in this particular experiment as a “carrier gas”? I’m assuming it is used for the GC, but is it more efficient than other gasses, or is it the only gas that won’t denature the extracted material?
I think that the degrading properties present in MSM is particularly interesting; slow degradation followed by a bout of rapid degradation seems to be something of importance.
Perhaps this type of degradation could be more beneficial in certain circumstances and for certain bacteria?
I’m also curious to know why the level of degradation is diminished with an increasing Fla concentration.
Although there is still successfully degradation, the relationship between residual Fla and the concentration of Fla is one that I would like to learn more about.
What about concentrations below 100 mg^-1? Would there be a faster degradation or maybe an unwanted/unexpected result?
Since the relatively low amount of degradation of Nap and Phe was contributed to “enrichment and continuous subculturing”, I am curious if there would be a way to remediate this by adapting the technique.
For example, what would the percentages of degradation of each respective PAH be if the enrichment and level of subculturing was altered?
Would these changes potentially enhance the PAH utilization capabilities of Rhodococcus sp. CMGCZ?
I think its so interesting how different strains here are more or less effective at degrading particular alkanes/aromatics, etc.
The last point mentioned about the degradation of diesel by these two bacterial strains makes me curious to know if there would be an impact by adding both strains together.
For example, would the treatment of both R. erythropolis T7-2 and Rhodococcus sp. strain Q15 in tandem with YE have an additive impact and therefore more drastically increase the level of degradation of the diesel fuel?
I’m curious to know the differences and similarities between Cycloclasticus and the Rhodococcus species that was investigated in the last paper.
For example, is the transformation and catalysis of a PAH by Cycloclasticus similar to the Rhodococcus process (are the ARHD’s the same, similar, or different)?
Also, I am curious to learn the specificity of catalysis by this bacterial strain. Is growth improved by the addition of YMSM? Are LMW PAH’s or HMW ones more efficiently degraded?
This is particularly interesting because it would insinuate that the marine bacteria have been able to develop pathways able to more effectively degrade PAHs in their specific habitats.
I’m actually surprised that degradation by marine bacteria has not been studied more being that PAH’s stand to cause harm to marine environments. So, I can definitely foresee this study providing unique and useful information regarding not only dangerous PAH’s in aquatic habitats, but also PAH’s in general.
The first thing that really caught my attention when reading this section is that it seems to be significantly more detailed when compared to the first paper. There is obviously more of a genetic focus in this paper, as evidenced by the ubiquitous amount of information relating to the sequencing and cloning. I’m curious as to why that is? Why was it more necessary to delve deeper with this paper?
This section of the paper really reminds me of organic lab, and I guess that’s justifiable. It is interesting to see overlaps between this course and others that I have taken.
One question that I do have, though, is what if some of these transformed products are lost? In organic lab, you may just have a low yield, but here, the products seem greatly more important. For example, what if excessive washing were to remove one of these converted products and therefore that information was entirely left out of final analysis?
I’m also interested in the relationship between the lac promoter and the expression of the dioxygenase genes.
My first thought when I read this section was that there shouldn’t be any associations between these systems because they seem to be unrelated.
I’m still wondering how the particular orientation of the PhnA constructs allow for proper expression under lac promoter control. Perhaps there are overlaps between the regulations of these genes?
It seems that the information provided at the end of this section insinuates the order of the phn genes is significant.
I wonder, since it was not discussed in the past paper, what the order of the phn genes were as it relates to the previous PAH degrader. How big a role does this order play in the ability to degrade PAHs? Is there a specific order that provides a greater degradation efficiency in certain environments?
I’m interested to know the details of why this particular gene possessed high homology with the Rieske center (and whether or not that is even relevant). I wonder whether or not there are other dioxygenases that are quite as effective as this one. Is the Rieske center constant among PAH degraders or are there others that have weaker, or perhaps stronger, impacts on degradation?
I notice that only in this phylogenetic tree is Rhodococci present. It’s interesting that a Rhodococcus does not exhibit the highest homology and there are also other strains that are very distantly homologous.
This really calls attention to the diversity of these dioxygenase genes and the differences between those bacteria that possess them.
I wonder then, since Cycloclasticus seems to be specialized for marine environments and Rhodococcus for terrestrial ones, if there are certain bacteria that are specialized for PAH degradation in other environments not yet explored in these first two papers.
This is interesting to me because it really shows the specificity of these dioxygenase genes. Obviously, there is something very different structurally existing in 3-Methylcatechol that allows for such a high level of activity by the enzyme here.
This then makes me question the evolutionary impacts that may have caused this. Perhaps this particular substrate was more common to come into contact with this enzyme and therefore it developed a higher specificity for it. I may also be looking too far into this…
A study that would look into the specificity of different types of dioxygenase genes would be extremely interesting. Although this study does touch on that, more information on the trends of these genes in different organisms may be able to give a more comprehensive outlook on their functions in different environments, in the presence of different substrates, and therefore their efficiency in different situations. Very enlightening paper.
I would be intrigued to know the evolutionary information behind this type of expression mechanism. Clearly it is extremely complex and very highly organized, so it would definitely be informative to know the role that evolution has played in this system. How fast were adaptations undergone in this system that made for the high level of adeptness we’re reading about? Also, are there potential ways for us to alter the expression of these genes to increase the efficiency of certain strains’ degradation capabilities?
I am curious to know the extent of involvement of the Archaean assemblage that the author(s) allude here. It seems that, as of now, their specific contribution is not quite elaborated upon. Are the Archaeans acting to actually form the biofilm itself? Or are they acting to augment the success of the arsenic metabolic activity? If they are impacting the metabolism, I wonder if this has evolved in this particular environment as a form of symbiosis? And if so, is it obligate or simply beneficial to the Archaean aggregate or the Bacterial bulk? This seems to be important information, but further reading may prove otherwise.
After reading through the paper, I’m wondering whether or not this type of metabolism with arsenic is ubiquitous… I understand that the author(s) refer to arsenic resistance as a “phenomenon”, but perhaps this type of unique metabolism has just not yet been characterized in depth. It makes me feel that certain microorganisms can use almost anything to their advantage if forced to, through evolution. Very amazing. Also, I wonder if there are other avenues through which this type of metabolism can succeed other than that of the biofilm?
One thing that stands out to me from the last paragraph is the differentiation made between the red and green colored springs. Here, it is stated that the samples were mostly taken from the red springs. I’m wondering whether or not this is just because the red springs are less rare, or if it has to do with the temperature…Or maybe they were the only ones with the arsenite traces and anoxic environments… I’m not sure if this is relevant but I’m curious why one was targeted over the other. Maybe those more rare green springs could have provided some interesting information to this study.
Another topic that made me curious about specific collection methods was the use of the “sterile toothbrush” for the slurry creation vs the use of the “sterile spatula” for the DNA analyses. The only thing I can think of here is that it is more important for the DNA sample to be more intact. But that makes me question why the sample taken for the slurry was not required to be fully intact/uniform. This, again, might be irrelevant…
This paragraph specifically mentions electron donor experiments, but never discusses electron acceptor experiments… Why is this? Would it just be redundant? Is all the valuable information discovered in the electron donor experiment?
What is meant by “killed control slurries”? It is the obvious answer that the organisms in the slurries have been… killed? Or is there something else important to recognize? And also, why is the autoclaving, which I am guessing did the “killing”, necessary to perform prior to the radioisotope amendment?
This section in a way relates to my first question in the introduction. I asked what role the Archaea play in this biofilm… It’s not a surprise that there is more diversity in the Archaeal groups because they are considered to be extremophiles. But I’m curious as to the extent of their contribution and the importance of their involvement.
The author(s) mentioning the possibility of an ineffective primer reminds me of the same problem that some of us ran into in our own testing. In this case, because they’re employing PCR, could this be due to the fact that there exists a new and unique gene that has not been previously characterized, as Dr. SNC referenced in the beginning of this section?
When I first read that there was an inability to discover related aoxB, my initial thought was that they must’ve found a novel system… I realized after that the more likely scenario was the primer incompatibility. So, I feel a little better knowing that it’s actual a possibility this mechanism could be something that has not yet to be discovered by anyone. Super interesting!
It’s really amazing to know that some of these extremely intricate metabolism mechanisms are almost as old as the planet. It really puts evolution into perspective… Some things have changed drastically while others have been maintained and stayed relatively the same. Our own evolutionary history is minuscule compared to this!
I think that they had a pretty good idea what was pushing the As(V) reduction, but they still needed to verify their ideas to be able to better understand the entire mechanism. This makes me so curious about how the system evolved and the types of adaptation that it had to go through in order to get to the point it is at today. Super interesting.
One question that I had throughout reading this (I also left some questions, too) was what is the exact role the archaean population playing in this system… It’s interesting that they actually have no involvement here because I expected their to be some kind of syntropy or symbiosis… It seems like they’re kind of just tagging along for their own benefit but they’re not providing anything of substance to the mechanism. Honestly a little surprised by this.
I too thought that the bioaugmentation was interesting. It was said to be the best method over biostimulation in the bioremediation of soils with low indigenous PAH-deegrading bacteria. This method goes against the Antarctic Treaty, which prevents the introduction of foreign organisms into the continent, unless it is used with native microbes. To get a better understanding on how bioaugmentation works, I found a simpler example of the method that used nicotine as the pollutant and synthetic tobacco wastewater as the medium for bioaugmentation. Tobacco is associated with the release of wastewater that contains toxic substances including nicotine. The synthetic tobacco wastewater showed a significant increase in nicotine removal.
This also stood out to me as well and I read that Antartica’s ecosystem is highly sensitive with pollutants and decomposition processes are extremely slow in this region. It’s very great to see a country that protects its environment and citizens from foreign substances.
This was a great point to bring attention to. I read that temperature is definitely a limiting factor in bioremediation. The temperature plays a major role in the rate and degree of microbial hydrocarbon biodegradation. This affects the viscosity of hydrocarbons. So, it is advantageous to increase the temperature when dealing with biodegradation in polar sites and this can be achieved by land farming.
Iron is an essential nutrient for growth and bacteria have to compete for the nutrient. This type of competition id indirect as it describes the rapid culture of a limiting resource. Siderophores come into play because they are used by bacteria to acquire iron. The competition comes because some bacteria are able to produce siderophores, while others are known as “cheaters” and only use siderophores but do not produce them. Species that produce siderophores with the highest affinity for iron has an competitive advantage.
Since V.harveyi cannot produce or import aerobactin, will this vibrio just be deleted from the study. Can it not act as “cheater” as the other vibrios?
After reading the title of this section, I really do not understand. Are they testing for chloramphenicol sensitivity to see if the vibrio is susceptible by the antibiotic or sensitive to it and is this apart of the mutant construction?
In the introduction, it said that aerobactin of V. fisheri prevents the growth of V.harveyi. Is this why the V.fisheri was chosen to become the mutagenesis. If not what is aerobactin?
This image was very important to the study because it set the tone for all other results. Here we see that the ES114 fluid contained an inhibitory substance that allowed for no growth. This is supportive of they hypothesis.
For this image, I was wondering in part A where they got the number five for five independent transposon from.
This is very important to understanding how and why V.fischeri produces aerobactin. That’s because it is needed for colonization of the v.fischeri’s host. The relationship between the host and itself is not harmful to either species, so it is more common that these relationships will form. It is just important for them to come together in the ocean that is sometimes depleted of iron.
Yes, this is interesting but I think they were saying repress iron uptake as in prevent iron uptake under oxidative stress conditions.
Nitrification promoting marine nitrogen loss are associated with the OMZs because they have high organic matter and remineralization. Just to be sure the remineralization is the process of nitrogen converting back to ammonium.
So because the Crenarchaeota are autotrophic, meaning they can consume inorganic substances like carbon dioxide, they are more important nitrifiers than abundant AOB?
Great point! I was wondering why Mn was included as they are indeed toxic to nitrification of bacteria. amoA gene would decrease because it is one of the genes encoding for the 1st step in nitrification.
Were the anammox bacteria already determined before it was verified by the CARC-Fish, and if so which step did this happen? I don’t understand how the bacteria was found to bring about nitrogen loss.
Is the Anammox bacteria and proteobacterial AOB abundance the same thing because if they were absorbed at the same depth then is there that much of a significance?
Yes, I feel this is correct Ryne the nutrient availability and oxygen content is the reason for the diversity. More life is able to withstand life here.
So, this “main objective” seems quite vague to me. I mean, I understand what they are saying, but “some naphthalene degrading bacteria” could mean 1 or 2 or all they find. I’d want to know which bacteria they were characterizing and why.
So, if these chemicals are harmful to mammals, would they not also be a threat to other species? Plants? Fish? Beneficial Algae?
It has always escaped me why scientists work on “should” assumptions. If the gene being studied “should” be critical, then why do we put the cart before the horse here instead of proving or disproving the function and overall dependency of the process of the gene?
How did they come to the conclusion that the bacteria was going to be a good candidate? They say it like it is a forgone conclusion, but that’s a pretty big assumption.
How was ZX4 chosen and why? What were it’s results on the initial phenanthrene growth?
Okay, I get that they added these chemicals to the medium, but what was the purpose of each? I notice that here it says the % of naphthalene degradation was calculated using this formula, but in another place it says it was “estimated.” Were these talking about two different things?
We have the shaker keeping the naphthalene oxygenated, but why would they not consider the effect that moving water or stable soil will have on the growth? This would definitely affect what is there.
So, as far as “biochemical” identification goes, how accurate is this? Of course I can’t name any off hand, but I’m sure that different species of bacteria can have some identical metabolic processes, can they not?
Haven’t ever heard of bootstrap replicates. What are these?
What about the benzene ring makes it more difficult to break down?
So, I understand that we want the oil degradation, and I consider this a good thing. But, what about the byproducts of the bacteria and their effect on the environment? We don’t want another dead zone like we have in the GOM though.
Just thinking about the big picture here…if the PAH’s are detrimental to mammals, why are we not also concerned with the effect the NDB will have on the Mammals and on the ecosystem as a whole? I mean, did we not just create a miles wide dead spot with the “cleanup” of the BP Oil Spill in the gulf?
Why are we only estimating the amount of naphthalene left? I googled it, and there seems to be several measuring methods for air, but not for water. Does the spectrometry not work for salt water samples?
In the introduction, they say that naphthalene and phenanthrene degrading bacteria have already been well characterized and that here we were looking at more diverse and quick adapting bacteria. Why then are we measuring degradation of phenanthrene here?
This is interesting, as I have read articles related to the effectiveness of bio-remediation using microbes to either absorb or degrade the oil in recent oil pollution. My question is, are there any long-term effects to introducing this organism to this environment?
The certain adapted bacteria inocula may be a key factor to discovering bigger solutions. I am curious to know the structure of the bacteria and how it bonds to each other, ie. does it create a net to degrade the PAHs?
Why were the isolates compared/controlled with E. Coli?
Would these isolates be tested for sustainability and use in bioremediation?
When using the KEGG model, does the morphology of the bacterium matter? Would the rod bacteria dominate?
It seems as if the P. Cirtonellolis would be an ideal material to use in bioremediation. It is important to consider the consequences that may arise when using PAH degraders.
Considering pyrene seems to be a difficult compound to degrade, BC1’s ability to do so is interesting. I am curious to know if BC1 is related to reputable PAH degraders.
“Bioemulsifiers act like surfactants but are more efficient in improving solubility…” This is the key to one of my previous questions. This is why BC1 was able to degrade the pyrene so well.
Using OD would not identify the effectiveness of the PAH.
I am curious as if this experiment used a control sample.
Does the darkness effect the success of the experiments? Also, what is the purpose of using KOH-soaked filter paper? Does the potassium act as a nutrient?
What are the benefits of selenium? Is it found in microbial mats?
The fnr gene is responsible for converting the selenium, producing red colonies. Its mutation is what disabled the E. cloacae from converting Se(VI) to Se(0). I had trouble understanding this originally.
I am still curious of the significance of the conversion of the Selenium. When thinking of the big picture, I understand we want to identify the genes and pinpoint mutations, but is there a significance to Se?
This concludes that the Enterobacteriaceae would be a potential bacteria for transforming selenium. I would be interested to see additional research to confirm this.
Because of the requirement of the fnr gene, the mutant E. cloacae is able to reduce Se(VI) through fermentation, which is an anaerobic pathway.
This gene diversity has to be in selenium’s favor for biodiversity.
Is the use of clones similar to creating a broth of the organism?
If selenium does not grow easily using the LB medium, why was it used?
Whats types of environments do naphthalene degrading bacteria thrive in the most and what types force them to compete with other microorganisms the most?
Does PAH bioremediation affect filter feeding or photosynthetic marine organisms such as phytoplankton/ zooplankton, corals, sponges, etc.?
Why were the 1000-ml bottles transported on ice?
How exactly is turbidity measured?
Why did only 18 strains show adequate growth?
Why was there such a dramatic decrease in growth after the 400ppm concentration?
What factors eliminated 6 of the 18 strains?
Could this study be done in other parts of the ocean?
I also agree that Madelyn’s definition of Bio-remediation was very helpful in understanding what this paragraph was trying to say. However, after just reading the articles explanation of it, in did not think it was safe. I did external research to find out that to complete bio-remediation they use natural biomes from soul and groundwater and just add more. My question is how, do they speed up reproduction in the organisms that are already there?
I believe that if bio-remediation does not remove PAH’s then it only does half of the job. Bio-remediation was said to be the most inexpensive way to do this, but it only handles degrading of components of crude oil. My question is what technique is in place to remove PAH’s and their fused rings?
What is the potential for pathogenic activity occurring in the in the marine population? These particular bacteria are pathogenic, how would overgrowth and infestation be controlled?
Studies show that the microorganisms are sensitive to pH changes. If the microorganisms involved in the bioremediation convert the PAHs to CO2 and water, how much CO2 is generated and what is the local effect of that increase on the microbes and or the local marine life?
What is the “growth and acidification of carbohydrates test?
What is the “growth and acidification of carbohydrates test and what does it tell us about the bacteria?
Would the sterile 0.2 micron filter paper be used to filter and hold the bacteria that may be in the seawater?
What is the benefit of using the “shaker incubator” instead of plate cultures? Does it increase aeration or is there another benefit?
Is the GC-FID measuring “growth” of the NP degrading bacteria, if so wouldn’t that destroy the bacteria, or is it measuring the naphthalene remaining in the media?
Has there been any research on higher molecular PAHs? Is there a real correlation between naphthalene degradation and their ability to degrade larger molecules?
What exactly in the structure of the gram-negative “outer membrane” makes it more suitable to degrade PAHs?
Is catechol production and the meta-cleavage pathway in all PAH degradation processes?
When DNA sequencing is done on the bacteria, is the plasmid sequenced as well?
What is the C230 gene sequence and what is the significance of it?
What is the benefit of the Biolog-GN microplates versus regular plates? Does it allow for a greater number of cultures or does give additional information than other types of plates?
Does transforming E. coli with the DNA of strain ZX4 to E. coli make it easier to observe the properties of the DNA encoding for the meta-cleaveage genes? Is this a common practice to transfer DNA instead of studying the isolated bacteria itself?
What is the significance in the decreased activity of C230?
Given the fatty acid composition of the cell, is there an advantage for the bacteria to have glycosphingolipids and no lipopolysaccharides?
Generally the closest genes will be transcribed together while others may or most likely will not. The genes phnH and phnI are only 19 bp apart and would be transcribed together. The phnG gene however, is 181 bp from the first gene and also is noted that there is a spacer and promoter sequences. This would prevent the transcription of phnG occuring with phnH and phnI.
Would the lack of similarity of GST in ZX4 to S. paucimibolis indicate a mutation in the bacteria? Is this related to the partial ORF for GST or is there a correlation between those two?
I believe the HMS (Hydroxymuconic-semialdehyde Hydrolase) is responsible for the step in the catechol, a substrate in the phenanthrene degradation. HMS hydrolases the 2-hydroxymuconic-semialdehyde by adding water and removing formic acid.
If the HSM was not available I would assume the pathway would stall and the degradation would not be completed.
Based on the information in this study as well as others that I have read, Sphingomonas paucimobilis appears to highly effective in the degradation of PAHs. In addition to the PAH degradation, Sphingomonas paucimobilis is also able to degrade xenobiotics.
The dissimilatory Arsenate reduction uses sulfide to reduce arsenate. In another paper I read, 1 mol of Sulfide oxidized to Sulfate would require 4 mols of Arsenate reduced to Arsenite.
Is sulfide the only inorganic anion used in this type of cycle?
Also in the other paper I read, the dissimilatory reaction was done in a curved, gram-negative bacteria also in the Mono Lake, California.
If the Arsenate is reduced to the Arsenite through this pathway, how is the Arsenite oxidized when it says it the biofilm is anaerobic and the photosynthesis is anoxygenic? Also they were no able to obtain PCR data for the Arsenite oxidase gene.
For the bacteria that have both oxidase and transporter genes, how does this affect the microbe’s ability to utilize arsenite or arsenate. I read another article that indicated that there was some resistance inferred if the microbe has both genes.
Are the singlet clones a product of the PCR/RFLP process or are they part of the RFLP probe?
From what I have read, there is a membrane respiratory arsenite oxidase (aoxB), a membrane respiratory arsenate reductase (arr), and cytoplasmic reductase (arsC). There is also a membrane transporter (arsB).
The arsenite oxidase oxidizes arsenite from the membrane as arsenite is used as electron donor on the cell membrane.
The arsenate reductase reduces the arsenate to arsenite again on the membrane.
If the arsenate is not reduced by the respiratory reductase in the membrane, arsenate may work its way into the cell. The arsenate molecule is very similar to the phosphate molecule and can enter the cell through the pit protein in the cell membrane. When this occurs, the cytoplasmic reductase then reduces arsenate to arsenite. I think arsenate has to be reduced in order for it to be removed from the cell. This is when the transporter comes in and removes the arsenite molecule to prevent cell toxicity with arsenite. Arsenite has a higher toxicity rate than arsenate.
There is a respiratory oxidase(aox) in the cell membrane that oxidizes the nitrite to nitrate on the in the cell membrane.
There is a respiratory reductase (arr)which is in the cell membrane and reduces arsenite to arsenate.
Neither of these processes has the arsenite or arsenate enter the cell.
Arsenate, being very similar to the phosphate molecule, can get into the cell using a phosphate transporter membrane protein. ONce in the cell, the cytoplasmic reductase (arsC) acts on the arsenate and reduces it to arsenite in order for the transporter protein in the membrane to export the arsenite out of the cell. Arsenite is much more toxic than arsenate.
Without the cytoplasmic reductase, the arsenate molecules would accumulate in the cell and kill it.
If I am understanding this correctly, in the anaerobic, dark environment, the arsenate did not reduce easily without out additional electron donors. With sulfide added arsenite was reduced to 1.75 mM. With H2, arsenate was completely reduced.
Does the concentration of sufide a 2mM affect the reduction? If there was a higher concentration of sulfide would it have been able to further reduce the arsenate?
If the bacteria activity was not changed by the addition of oxygen, would that show they are not using water for electrons or oxygen for the electron acceptor?
H2 appears to be the best electron donor in the dark reactions. The redox reactions were much faster with the additions of H2 than with H2S. Incorporation of C14 was higher with H2 as well. Although there is no test done with H2S, I would think the incorporation rate of C14 would be lower than H2 as is the reaction rate.
What would cause a bacteria to use the arrA gene versus ars B gene? Would it be driven by an overwhelming concentration of arsenic?
Or is it triggered if arsenate is able to enter the cell and then the ars C gene reduces it to arsenite and ars B just clears it from the cell? Is this just another reduction process?
Is fluoranthene easier to degrade with the relative less stable configuration than the alternant PAHs?
Some articles I reviewed indicated that HMW PAHs, that were not degradable or poorly degraded, were successfully degraded, such as fluoranthene, when cultured with phenanthrene.
Is the phn a method for genetic transfer? I was trying to read about the genomic islands and phn and I saw some different GEIs such as PAIs for pathogenicity island but was not certain what the phn is. I thought maybe it’s phn for phenanthrene degradation.
In curiosity, I looked for the composition of the 216L agar. I found this description of this agar in this article: Yuanyuan Fu,1 3 Xixiang Tang,1 3 Qiliang Lai,1 Chunhua Zhang,2 Huanzi Zhong,1 Weiwei Li,1 Yuhui Liu,1 Liang Chen,1,3 Fengqin Sun1 and Zongze Shao. 2011. Flavobacterium beibuense sp. nov., isolated from marine sediment. International Journal of Systematic and Evolutionary Microbiology. y (2011), 61, 205–209.. The agar is composed of: sodium acetate, 1.0 g; tryptone, 10.0 g; yeast extract, 2.0 g; sodium citrate, 0.5 g; NH4NO3, 0.2 g; seawater, 1 l; pH 7.5. What classification of agar would this be? I was thinking enriched?
Is the Cre-lox recombination method a knock-out process? If I understand the process, the Cre recombinase will cut a section of the DNA out, invert or transpose it. The lox is a marker that is on either side of the target.
When the dioxygenase is knocked out, does that remove both the alpha and beta sub-units go together?
When you knock-out a gene of interest is it possible to remove only the gene of interest does it remove a cluster of close genes?
In comparing the P73T genome with the C. baekdonensis
B30 genome, it is curious that they are similar in genetic material but the functions are different. With the genome similarities would it be possible that the PAH degradation genes are foreign and on plasmids?
Also in another section (paragraph 35) it states that the B30 region was unable to degrade PAHs but in this paragraph states “only 3 RHDs were identified in the B30 region”, if there are 3 RHDs I would think it could degrade something.
Thanks, I was so focused on the PAH I didn’t even think they’re not the only aromatics.
I may be just confused but, in paragraph 2 the report is there were numerous hydroxylating dioxygenases and ring cleaving dioxygenases for PAH degradation and then in paragraph 4 it is reported that the C-7,8 dioxygenation pathway is the only one. Did they find potential genes but expression of only one pathway?
This appears to be the most interesting, experimentally, of the bacterium that we have read about so far.
This is very interesting and highlights that in nature there is no one reaction. It is interesting to me that they are researching foreign bacteria for the bioremediation. Wouldn’t it not be reasonable to isolate native bacteria?
I would have thought there would be bacteria capable of degrading the PAHs in the area and some that would also have acquired heavy metal tolerance.
Symbiosis at work!! If my impression is correct, the ryegrass is able to degrade, or remove or just hold on to the heavy metals which decrease the efficacy of the degradation enzymes so the bacteria are able to degrade the PAHs.
With the insolubility of PHE in water, methanol is used to dissolve PHE for inoculation in the soil. Then CUCO3 is added to the soil. What is the significance of deionized water?
Does methylene chloride react with CuCO3 in the extraction of PHE?
Pertaining to the abiotic factors:
A change in the natural environment that would reduce the bacterial population thus reducing the degradation activity such as pH changes or moisture. I read some information that chemical oxidation, potassium permanganate, reduces PAH concentration minimally. The decrease PAH concetration is minimal but higher degradation amounts were achieved with a pH 7.5-8.
But I would think those would occur naturally and not in a controlled environment??
This paper is much more interesting. Going through some of the experiments at the same time as the paper makes it easier to put the pieces together.
Copper is a trace element in plants as well as humans. Copper is involved in defense responses in plants and enhances some plant enzymes. It kind of makes sense that, to an extent, copper would enhance plant activities which in turn assists the bacteria by nutrients needed. I had also read that copper is also directly affecting the dioxygenase activity. Is that correct?
I thought the heavier metals such as iron generally inhibit the degrading enzymes. After reading this through again, I am not sure about the last statement. Other than a toxic level of copper would it replace iron in the enzyme. I thought copper is less reactive.
Is there a relationship between the concentration of PHE and Cu that increases a higher Cu concentration for the plant? Cu does play a role in enzymatic activity but I I can’t find a direct answer that relates increase uptake of copper by the plant. Unless it is just the cycle.?
Yes it is the same process however it is done under nucleotide blast instead of the BlastP that we ran for the RHD genes. The Mega process would be the same except is again is nucleotide not protein.
Could the inability to identify the gene because of non-matching primers? Could there be a missing “catalyst/inducer ” that initiated the reaction or improved the process ?
I may have a one track mind but is there something missing if it has the genes previously proven to degrade but it can not metabolize fluorene?
Or is the pCAR3 foreign to the host and is unable to express or,again, needs a little help to function?
Is the third degradation pathway very likely to occur? It appears to be a less favorable pathway to me. This reaction would be from a mono-oxygenase instead of the same dioxygenase as the other two pathways?
I read that there was an increase in secondary structures or mismatch in primers with multiple repeats of runs. Is that correct?
What exactly is an angular dioxygenase? Does it have to do with a different attack site?
Is there a benefit to using the phosphate buffer? I read that it the Tris buffer is used during enzymatic processes and nucleic extraction. Phosphate buffers are not affected by change in temperature but Tris is.
Primer walking uses digests of DNA to sequence the genome. A primer starts a strand of DNA and that fragment is sequenced by chain termination(Sanger). Then the next primer used is formed from the end of the sequenced strand before. The process would continue until the genome is sequenced. If I am understanding this correctly.
Why is flnA1-flnA2 not able to degrade Carbazole? If I understand this correctly, Carbazole has a hydroxyl added then flnA1-flnA2 acts on it and becomes a dihydrodiol and flnA1-flnA2 changes to a linear action instead angular??
The flnA1-flnA2 dioxygenase complex is able to produce the three oxidation products listed in Table 2. DFDO is able to only produce 1-Hydro-1,1a dihydroxy-9-fluorenoned and CARDO is only able to produce Dihydroxyfluorene with Fluorene as the substrate and neither of these produced Fluorenol-dihydrodiol. Is this a specificity or approach different in the three dioxygenases?
I think what they are saying is that the majority of studies with angular dioxygenases have been in gram positive bacteria and the primers tested were from gram positive bacteria. Perhaps that contributed to the lack of amplification.
The granule version of Carbofuran was in fact banned in 1994. It was banned for bird deaths from ingesting the granules as well as the deaths of animals that ate the birds contaminated with the pesticide
The mcd is a gene encoding a metallohydrolase for the initial hydrolysis step in the degradation of carbofuran.
This paper is intriguing. I appreciate the difference in the subject matter, meaning this is an intentional act to “protect” crops that may actually be causing greater consequences.
I am having some confusion in this paragraph. Is the identification of the metabolites of Carbofuran identified by comparing to known pathway metabolites using the processes listed?
In this situation, these are bacterial conjugations used to randomly transfer the Tn5 transposase gene and Km resistance to construct a mutant library.
Would the loss of the expression of the gene cluster cfdABCDEFGH be because the CftA gene is a transporter and without the transportation there is no need for oxygenase expression?
In reading the Fig 3, It looks to me as though all the mutants as well as KN65.2 can complete the initial step,as the concentration of Carbofuran decreased from 0.8 to 0 either immediately or within 2 days.
In this paragraph, the mutants in group V are not able to degrade the carbofuran completely thus accumulated carbofuran phenol. The mutation of cftA not only interrupted the ability of carbofuran into the cell but also decreased the expression of the gene cluster believed to degrade carbofuran. As the accumulation of the metabolite 1, carbofuran phenol, increased, an alternate transporter gene was strongly increased to remove the toxic build up. Am I interpreting this correctly?
Would this expression similar to lac operon minimal expression? Or could it be expressed for a different purpose? Aren’t some of these genes involved in some type of fatty acid synthesis? (Making enzymes that aid in the degradation).
Very difficult paper for me. I am struggling to put all of the parts in a organized and concise manner.
Hey Julie,
I too am fascinated by Rhodococcus and its ability to transform harmful PAHs into a TCA cycle.I believe it has a promising future for the clean up of polluted areas. Also as you’ve listed in paragraph 2, “PAHs are found in coal, wood, and other materials.” The Rhodococcus strains are in fact efficient at removing hazardous elements such as sulfur from coal and petroleum that are found in PAHs, thus benefiting our well-being.
I thought that Rhodococcus belongs to the family of enzymes known as ARHDs which should have exhibit homolgy with catalytic domain even for previous species? Could there been a change in its bioremediation? Did bacterial evolution change its functions over time?
The thin layer chromatography was a fun experiment somewhat similar to streaking in my opinion. Analyzing the spread of both and the migration of the spread over time, also being careful not to rip through the surface while doing these experiments.
Could colonies grow even after a certain period of time? Also Does MSM and R2A plates form different numbers of colonies or does it depend on how well you perform the experiments?
Did they have various degradation test for MSM with the concentrations of Fla? How was the Fla degradation percentage calculated in those eight days?
Knowing what I know now about MSM and R2A plates, Im definitely getting more comfortable with the lab preparations such as being able to streak and being able to describe the colonies just by looking at them. For example the different colors blue or pink, characteristics in the colonies such as pure culture and observing the growth of them with time.
The R. erythropolis in YMSM in figure 2 shows on day 1 to 3 the residual Fla percentage dramatically dropped about 60 percent while the R. erythropolis in MSM shows a slight drop. Gradually over time the MSM also had a huge drop in percentage from day 5-7. As for YMSM there was just a constant percentage drop.
hey anna , I also think its really cool and fascinating that Rhodococcus species come from a diverse tree with mycobacterium being the best relevant match. Also the tools to figure out the closely related, its specific gene, and the number of nucleotides. Maybe its possible that they have the same functions because they do seem to have very similar attributes.
I read more great things about Rhodococcus with its unique functions and diverse abilities that make them such a special case. Im glad that there are studies in pharmaceuticals and chemical industries that will help benefit the environment. Im hoping that Rhodococcus will become a well known compound that performs efficiently in many diverse roles in the near future.
How efficient is the tool to amplify “bp” measurements? Also in this case does Rhodoccus sp.CMGCZ expected to not go through with its regular degradation with a different bacterial strain PAH dioxgenase gene due to bp? How close does the bp band have to be for regular degradation to happen?
I find it fascinating that Cycloclasticus has been reported as another bacteria that is able to degrade PAHs in marine environments. I was in belief that Rhodococcus was only the bacteria to degrade PAHs, I’m interested to see the breakdown of this bacteria and to see if there is any similar functions or characteristics as Rhodococcus.
Im wondering what the level of toxicity is for PAHs? For example when dealing with burning of grass fires or even smoking cigarettes, do you immediately react to the exposure of PAHs or is it long term effect? Also now there is a possibility that seafood consumption is contaminated. Hopefully Cycloclasticus is capable of clearing the contamination in marine environments.
Im glad that the chemicals used in the laboratory is highest purity because they will be able to obtain the highest quality and purity of reagents.
Im wondering if sonication was purposely used in the experiment. Was this effect used to open the cell for further use and the result was supernatant as the crude cell extract?
It’s interesting that phnA(1-4), phnC, and phnD genes are key players for the degradation of PAHs. I notice that the organism Sphingomans sp. exhibited the highest percentage of identity labeled on Table.3, also showed the strains had the best sequence with the amino acid of ISP subunits of aromatic oxygenesases. Im wondering if these genes are effective to the degradation of PAHs.
Ive noticed that phnC reveal highly significant percentage then any other gene. It’s possible that phnC has the most potential for example, high activity with different substrates. Im interested to see how well does phnC contribute to the Cycloclasticus in degrading PAH.
After reading paper 2, I believe the phnC gene is the best candidate for bioremediation because it exhibits 60-66 % identity with amino acid sequence(highest group) and can degrade PAH dioxygenase efficiently.
Im still wondering if these arrows indicate splicing or is it a form of transcription/translation?
Its interesting that PhnC gene encodes the PAH extradiol dioxygenase with substituted monocyclic catechol compounds. Also its involvment of both upper and lower pathways for degradation of napthalene, phenanthrene, and biphenyl. Im hoping researcher will find the true substrate of PhnC that gives them a better insight on the pathways of degradation in Cycloclasticus.
If phnA enzyme could not be used in anthracene as a substrate does this mean that E.coli has the same characteristic?
Im surprised that some microorganisms are able to withstand normal existence when high concentrations of As(III) and As(V) are present. Im excited to see the results in oxic and anoxic, however I believe researchers are focusing arsenic under anoxic conditions because it’s only dependent on the amount of organic matter. It may be predominantly better assuming the results will be more efficient. -I may be wrong
It’s interesting that the light driven from anaerobic ecosystem may have played in the Archean Earth. It’s possible that this anoxic environment used light such as redox couples As(III) and As(V) serving as electron donor and acceptor for obtaining energy to survive and later evolved.
Its interesting that slurry mixture were stored and preserved in the dark for several months prior to the experiment. My question is whats the reason for a mixture to be left for so long?
The light and dark incubation having an effect on the electron acceptor and donor of an arsenic. Does this change the integrity and activity of a bacterium? For example the photosynthetic bacterium Ectothiorhodospira strain PHS-1, can it evolve quicker under light with As(III) condition vice versa of a different situation using dark.
It is just to try and further prove that AS(III) oxidation and As(V) reduction can happen together.
In order for bacteria to thrive in harsh environments I would assume that they would have similar structures to Archaea since Archaea are known to live in harsh environments.
I would assume that the explanation dealing with the primers being not suited for the environment and the solution being to try a new primer is more plausible than it being a new microbe.
It’s interesting that the microbial populations were more effective in light incubation expressing similar pure culture.
Im wondering if dark incubations had a specific temperature will it change the process of biofilms.
Its interesting to see that As(V) reduction could be viewed as an ancient phenomenon that dates back 3.8 billion years ago. I actually would have loved to see more about the evolution and phylogenetic diversity.
How does the co-substrate yeast enhance degradation?
Since the structure of PAHs contributes to their biodegradation resistance, how does degradation take place in PAHs? Does the fused ring break apart first?
Anna, I am also curious as to why the phylogenetic trees were constructed. Were the phylogenies used to determine which bacteria was a more efficient PAH degrader? Or did the phylogenetic tress help the researcher have a better understanding of how each bacteria played a role in degradation?
Why was the temperature taken at a depth of only 1cm? Would taking the temperature deeper into the water yield different results? I am curious about how the temperature of the soil sample affects PAH degraders.
In locations B and C, the concentration of extractable PAHs and the ionic concentrations are high. Locations B and C had higher ionic concentrations than location A. Since these locations are rich in nutrients required for bioremediation, would there be a higher success of natural bioremediation in locations B and C than location A? Or would the success rate be nearly the same due to low nitrogen and phosphorus levels and the high PAH concentration?
Since nutrient-rich media favor rod bacteria over coccoid bacteria, would we only see rod bacteria in the different morphologies? The text stated that rod bacteria can out-compete cocci bacteria due to their surface contact. If both bacteria are together, would rod bacteria take over completely? Or would a small amount of cocci bacteria be present with the rod bacteria?
Anna, I struggled to understand this section, but after reading your comment about using this data to construct the phylogenetic tree, this makes sense. I think that creating the phylogenetic tree helped in the identification process. This section outlines the similarities that the isolates have to classes under the Proteobacteria phylum. The next paragraph explains the similarities in the isolates and the classes, as well as degradation processes within the classes. I agree, the construction of the phylogenetic tree does lead you to think that related bacteria can be useful PAH degraders.
The text states that there is almost no record of the use of P. citronellolis in PAH biodegradation. Since it is not specified as a pathogen, and it had an astounding amount of growth on the PAH substrate, why haven’t more experiments been performed with P. citronellolis? It seems as if P. citronellolis would be very beneficial in PAH biodegradation.
I am a little confused on how the hydrocarbons are recycled. Do the vent fluids bring the petroleum hydrocarbons upward from the deep sediments? Or are the petrochemicals within the vent fluids responsible for the hydrocarbon recycling?
The text Staes that not much has been published in regards to deep water hydrocarbon degradation. Could hydrocarbon-degrading bacteria present in coastal waters also be beneficial in deep waters? Could the same bacteria perform degradation in deeper waters, or does the depth and temperature of the water play a role?
I am also interested to see if each sample contains unique PAH degrading bacteria. Core 4567 was collected from a deep water environment, with no sulfide, and no bacterial mat. Core 4571 was collected from a deep water site, but it was oil-abundant with a bacterial mat present. Core 4571 also had sulfide present. Since each cores were collected from sites with very different conditions, I would expect the PAH degrading bacteria to have different characteristics regarding to where it was collected.
I am unsure of how the aluminum foil prevents hydrocarbon sorption. Does it serve as an extra protective barrier around the cap? Another thing that stuck out to me was that all of the flasks were incubated on the orbital shaker in the dark. Is there a reason they were incubated in the dark?
By looking at the graph provided, it seems as if there are drastic differences in the amount of 14CO2 recovery from each sample. Compared to core 4571-2 (solid bar), core 4567-24 (open bar) seemed to have a great amount of 14CO2 recovered for NAP,PHE, ANT, FLU, and PYR. Core 4571-2 (solid bar) had a higher amount of 14CO2 recovered for BaA, but it was not as drastic as the others.
The study found that Cycloclasticus could be a major hydrocarbon degrader at the Guaymas Basin. If Cycloclasticus co-exsists with other hydrocarbon degraders in surface sediments, what makes it have poor cultivability and hard to isolate compared to the other hydrocarbon degraders?
Performing microbial bioremediation is particularly difficult in Antarctica due to the extreme climate and constant low temperatures. Microbial bioremediation processes are slower, compared to the rate if administered in warmer or tropical regions. It also affects hydrophobic contaminants in the region such as the PAH from diesel oil. Hydrophobic (lipophilic) contaminants tend to be more durable and more difficult to remediate in low temperature regions due to the slower biodegradation rate.
– I was not familiar with the term anthropogenic pollutants or bioaugmentation as a remediation strategy for pollutants. An anthropogenic pollutant means that the source of a pollutant is a direct result of human activity. Bioaugmentation is a microbial bioremediation process that works by adding a specifically isolated hydrocarbon-degrading bacteria that have been cultured in the lab (ex-situ). Indigenous Antarctic hydrocarbon-degrading microorganisms are often the best adapted to the environmental conditions. For this reason, they are a good candidate for bioremediation/contaminant cleanup from oil spills.
The multi-process technique for phenanthrene quantification caught my attention, and I did some further research. It seems that every variable in this study is substantially complex and highly specific. In this case for quantifying three-ring PAH concentrations, the combination of excitation-emission fluorescence spectroscopy and multivariate data analysis are useful for modeling degradation studies and monitoring PAH concentrations and microbial growth under Arctic conditions. Recent technological development now utilizes multi-dimensional EEMs, derived from EEM fluorescence spectroscopy, scan excitation and emission ranges simultaneously, reducing broad spectra, and increasing selectivity.
The aromatic ring structures within the PAHs and their hydroxylated metabolites possess naturally fluorescing properties. Fluorescence spectroscopy may be used to analyze the concentrations of these compounds to determine degradation and metabolite production. With multiway data and PARAFAC analysis it is shown that reliable concentration determinations can be achieved with minimal standards in spite of the large convoluting fluorescence background signal. Thus, rapid fieldable EEM analyses may prove to be a good screening method for tracking pollutants and prioritizing sampling and analysis.
Microbial remediation is not solely effective in cold regions, but is the most suited technique for harsh, arctic conditions. In cold environments, oil-based contaminants remain longer than in temperate regions due to the low bioavailability of hydrocarbons and the harsh climate.
Natural remediation by Antarctic soil microorganisms is slow due to low availability of liquid water, lack of nutrients to support microbial growth and low temperatures that reduce bacterial metabolic rates. One particular bioremediation strategy suited for cold regions operated by introducing native Antarctic hydrocarbon-degrading microorganisms, as such organisms are often the best adapted to the specific environmental conditions.
Oil spills generally have a negative impact on soil biodiversity, in which overall microbial biodiversity may decline.
Following an oil spill, hydrocarbon-degrading bacteria can increase significantly in number, and may become a major proportion of the total culturable microbial population. This study confirmed that diesel exposure has favored the development of microorganisms that use hydrocarbons as a source of energy. Antarctica’s microbial communities inhabiting regions near an oil spill are excessively exposed to hydrocarbons, which then caused a selection for microbes that are capable of utilizing PAH as carbon and energy source.
Bacterial staining and dye techniques are fascinating and gives me an appreciation for how far science and microbiology has come. The importance of bacterial detection on agar plates is paramount and will indubiously continue to evolve as microbiological techniques advance.
I agree with you, Julie. I believe that the exploitation of rhodococci and their unique ability to transform PAHs into different intermediates can greatly benefit human society and health, as well as the atmosphere. The use of rhodococci could significantly lower the risks of human diseases by removing hazardous environmental toxins.
I predict so as soon as scientists can hypothesize a method to harness the abilities of non sporulating aerobic rhodococci. It is important that more studies such as this are done in order to prevent further pollution and toxic compounds from entering into the atmosphere.
I am interested in this as well. I wonder if there would be cases of negative results during microbial manipulation due to the addition of organic/inorganic substances. I suppose it would all depend on the microorganism isolated.
I was thinking the same when I read about the use of the minimal salt medium. Since it seems to be a commonly used practice in laboratories, I am interested to see how effective the MSM is at isolating pure cultures of bacterial strains for our labs.
I am finding many similarities between the scientists’ methods and procedures and the ones we have used in our lab 2. The spreading of diluted cultures on agar plates seems to have yielded pure cultures of bacteria. I am excited to see how our bacterial children grow and develop after being incubated.
It appears that the bacterial strains had a high affinity for Fla compared to Nap and Phe. The difference in percentages of the PAHs degraded varied greatly between Nap/Phe and Fla. I wonder why in the presence of YE that the strains were not capable of degrading Nap, yet Phe and Fla were still able to be degraded.
I wondered this as well and what made the bacterial strains have such an affinity for Fla compared to Nap and Phe. It probably has to do with the culture mediums on the agar plates, but I am still curious whether there is a way to better select and isolate a substance for degradation.
I am curious to know what kind of pharmaceutical and biotechnological advances we will be able to make with Rhodococcus. Manipulating and exploiting their metabolic abilities will continue to progress modern technology. The use of their enzymatic properties will perhaps give us more insight into the nature of bacterial strains.
Rhodoccocus preferred Fla as a source of carbon over Nap or Phe given that it degraded almost 100% of the Fla and small percentages of Nap and Phe. I wondered why this was after reading the results last time but understand now that it was due to the enrichment and subculturing techniques these scientists used.
I assume that obligate marine bacteria are more capable of degrading PAHs than terrestrial bacterial strains due to the presence of excess pollutants in seawater.
I am interested to see how the Cycloclasticus strain of bacteria compares to the previous PAH degrading Rhodoccus bacteria in the last study. It seems that it will have more of a capability to degrade oils than did Rhodococcus.
I am interested to know more about how the lac operon was used in this experiment and how it affected the results. With the ability of the lac operon to repress or activate cellular activity, I suppose there are numerous outcomes during experimentation depending on what genes are manipulated.
This interests me because I began writing the pre lab for Wednesday and I recall some of these procedures in the protocol. I am excited to be able to use these techniques on our microbes. Centrifugation is fascinating in that it separates cells into layers due to high speed spinning so that you can visually see the different components.
This passage interests me even more after it was explained in class Wednesday. I feel I have a better understanding in these methods as far as using plasmids as vectors.
The ability of these strains to turn the indole to indigo is very similar to the microbes we are working with in lab. This makes me curious about whether our bacteria contains aromatic oxygenase genes.
I, too, find it incredible that we are able to work with genetic sequencing to determine the chemical abilities of microbes. I am also interested in the Cystoclasticus strain and its PAH degradation abilities compared to the strains we have been using in lab.
It is interesting that the PAH degradation genes were located on plasmids in Pseudomonas strains but on a chromosome in the Cyclocasticus species, yet both genes function similarly despite being on different genetic locations.
I am interested to know if further studies will be made involving the efficiency of the other dioxygenase genes of Cycloclasticus. Since it has been established that two different species of bacterial strains that can degrade PAHs have been discovered, I assume there must be many more bacterial processes that we can use to our advantage in industry.
I am wondering the same thing because I thought the addition of more PAH degradation genes would increase the degradation potential of the bacteria.
I was also wondering this as I was unaware that it was possible that a bacterial strain was unable to produce a usable PCR product, as this possibility has not been mentioned. I wonder if it has something to do with the activity of the primer or polymerase.
I am actually surprised that bacteria can exist in environments rich in arsenic, arsenate and arsenite considering the level of chemical extremity. I was under the impression that archaea would have been the only dominating prokaryotic organism in these environs.
I find it fascinating that this study will be involving Archaea, as it is incredibly interesting that these microoganisms can live in such extreme environs such as hot springs and regions high in volcanic gases. I am excited to see what these scientists uncover as the experiment progresses.
I am curious to know why the samples were stored in the dark and why this was important to the experiment. I was unaware that that would have an affect on isolates.
I am interested to know why the dark incubated control groups were wrapped with aluminum foil and what the foil’s function is for the purpose of this experiment. Is this just to shield the sample from the light or is there another purpose?
I am interested in knowing more about this particular phylum of Archaea and its affinity for high salt environments.
It is incredibly interesting that similar strains from the same red biofilm had vastly different temperature ranges, yet their optimal growth temperature was comparable. This makes me wonder about the differences in their metabolic functions.
I wonder if the use of different kinds of primers suitable for this particular environment would have resulted in better results in regards to the detection of arsenate oxidase genes.
Would this be an example of when horizontal gene transfer/mutations within genomes of bacterial species complicates 16S rRNA phylogentic analysis?
What I gathered from this paragraph is that coastal spills are the most detrimental pollutants to the environment, but there are safe ways we could improve and handle this. I was still a little confused by the concept of bioremediation so I googled the definition which made it much more clear so I could move on to the next paragraph. For those as confused as me… “Bioremediation: process used to treat contaminated media by altering environmental conditions to stimulate growth of microorganisms and degrade the target pollutants”
So if I read this one correctly, is it now saying that bioremediation is not efficient at removing PAH’s which is what they are about to try to do? My next thought is, there has to be a something that can break down the fused rings of the PAHs to make them less resistant.
So now there is reported success with bioremediation used to degrade PAHs, however the trials include co-substrates that help make it successful. They are testing it with no added substrates to see how much the co-substrates were taking credit for in degradation. From the 4 paragraphs i’ve read, my first thought is that the co-substrates helped a lot.
It says that the temperature was taken at a depth of 1cm. Was there any way that the water affected the thermometers readings considering 1cm is not that deep? Is that not just the same as taking the waters temperature? I am confused what taking the temperature was actually for.
Does suspending the pellets in BH medium make a concentrated bacterial solution because the pellets dissolve or because they just contaminate the BH medium? I do not see anything else mentioned about the pellets so I am assuming they dissolved and formed the later BH liquid?
Not a question. Just pointing out that an increase in microbial population results in a clearer culture solution. This stood out to me because I though the opposite would happen until it said that the weight of the bacterial cells makes them settle to the bottom, which then made sense.
So I thought gram-negative bacteria would appear pink in color. Is this only on certain agars? It says these colonies were cream and clear but they are gram-negative. Maybe I am getting something confused?
I found it interesting as well that the rod shaped bacteria can out-compete cocci. It makes sense that they have better contact with the surface, it just surprised me that they were that much more dominant.
No questions. I just took away that natural bioremediation has many studies in deep petroleum reservoirs and coastal environments but very few in deep-sea waters. Since more exploration is happening for oil in the deep-sea, it makes since to learn more about natural bacteria that could degrade any oil mess made.
It seems like deep-sea degrading bacteria live at high temps. This makes me think that they are a different strand or type of bacteria than the shallower coastal water degraders.
I don’t think the -80C will kill the bacteria. We are storing our bacteria in lab this week at that temperature to make the freeze stock of it. Based on this saying “prior to storage”, I imagine they are doing something similar to us storing ours.
So they inoculated vials with the wet sample that was collected… but it says they prepared the killed controls prior to inoculation. What killed controls?Also, it says all treatments were done in triplicate… Is that saying that there were three samples of each of PHE, ANT, PYR, FLU, NAP, and BaA?
Okay so i am confused on how many flasks there are… is it saying they had 15 flasks containing 18 ml of ONR7a medium and 2 g of sediment slurry from core 4567-24 and then they took 5 of them and added 1 mg of [U-13C]-PHE and another 5 and added 1 mg of unlabeled PHE??? I hope this makes sense because I am confused on what they are replicating and duplicating
PCR makes many copies of DNA whereas qPCR targets a DNA molecule during PCR. So i’m guessing they used qPCR since they want to know the amount of the specific OTUs?
They were successful at identifying PAH degraders in the deep-sea in the Guaymas Basin sediments. So what is next? If these PAH degraders exist, would they not already be showing a significant influence on the present crude oil?
Am I reading this correctly… Are they saying they are trying to see if the same Cycloclasticus PAH degrader found in shallow water sediment is also found in deep-sea sediment along with other PAH degraders?
I feel like this paragraph can easily be shorted down for the summary. The main take away seems to be that PHE showed the highest mineralization so they used it to continue on in the experiment.
Im guessing that the main take away here is just the process of tracking C13 with the use of C12 and C14. I wouldnt think knowing that it took 11 days matters for a summary.
So toxic states of selenium (selenate & selenite) can be reduced to selenium with the help of microorganisms. Bacteria that can respire Se use selenate and selenite as electron acceptors and then emit selenium. Even those these bacteria are very common, we still know little about what they are capable of.
Each Se-respiring bacteria emit selenium but the structure of the selenium is different depending on what bacteria strand it comes from?
Im assuming we are about to try and isolate selenate reductase genes from microorganisms.
1.) The cells were grown in Pyrex bottles in LB Broth 2.) Cell density and oxygen concentration in the pyrex bottles was monitored3.) Cell were harvested from the pyrex bottles and suspended in salt medium4.)Antibiotics were added to the cells in the salt mediumAt what point were the strains grown on LB agar?
The first 5 sentences seem very detailed… is our main takeaway from this paragraph suppose to be that three strains were tested for selenate reductase activity based on the color change on the agar plates?
Okay so E. coli S17-1 was not able to reduce Se(VI), but E. coli S17-1 combined with cosmid clone E. coli pECL1e was able to reduce Se(VI)?
So the nucleotide sequence of 9.75-kb DNA fragment was determined… it had 8 ORFs… it was then subcloned into 4 fragments…then what is it saying? They looked at these 4 fragments to see if complete ORFs had fnr or ogt genes so they would know which one was responsible for selenate reductase?
So they all came to the conclusion that E. cloacae cant use Se(IV) as its only electron acceptor for anaerobic growth? They are comparing the current experiment with previous studies, right?
So FNR regulation is active without oxygen present… so when there is no oxygen the fnr gene is expressed and that activates Se(VI) reduction. Is that correct?
So here they are saying E. coli doesn’t have the oxygen sensing transcription factors so it doesn’t know if oxygen is present or absent, therefore it cant reduce Se(VI) because the reduction of Se(VI) happens due to FNR regulation that starts in the absence of oxygen… right?
Okay so E. coli does reduce Se(VI), but it is not from FNR regulation activating it. Rather it is from the presence of the YgfK, YgfM, and YgfN proteins. So is this part just saying there are multiple ways to reduce Se(VI)? This discussion section is confusing me.
I am curious why we haven’t seen this as frequently in marine bacteria- is it because they are less studied in general or they have been studied but this mechanism is still not seen? This would be an interesting topic to study among marine bacteria as they could target the marine pollutants as well.
Given the differing environments, I also think this could be a semi-universal gene/genomic island but it is only expressed in certain environments or in differing conditions. It also makes me wonder about post translational modifications or epigenetic factors that could affect the protein expression.
I am looking forward to their methods and results because I wonder if this wide range of degradation is due to one extensive metabolic degradation pathway or several unique pathways. Given that they are looking into the first fluoranthene degrading bacterium, I would think it is a specific pathway but I would not be surprised to see familiar intermediates and mechanisms. In breaking down the fluoranthene, are there harmful byproducts or anything the bacterium can use for its own benefit?
If this was the only mutant strain, did they have/need a positive or negative control?
I wonder why they had to predict the remaining protein coding sequences? Were their samples degraded or not fully manageable due to their environment? Depending on the accuracy of this technology, this could affect their results greatly.
I am most familiar with vectors inserting a novel gene to a plasmid, but it appears this vector carries ampicillin resistance and B. subtilis synthase subunit. I am not sure of the significance of the latter but it appears this is also introducing the deletion of the target, knockout gene. I am looking forward to their results and discussion so that this will all start to come together more.
This is very interesting that the P73 strain was able to acquire the PAH degradation properties from a bacteria in another order via horizontal gene transfer!
It is interesting that the most HTG genes were related to the rhizobiales but the greatest overlap in the proteome was directed toward an unrelated strain, C. baekdonensis.
This is really useful results of the study! By narrowing this down, they are able to determine which other genes could be necessary for PAH degradation.
Further research could confirm this with a gain of function mutant study to further observe the pathway in action.
I agree, this is a great, defining statement reporting their unique results. It is incredible that they found a protein that is a part of the most difficult step of fluoranthene degradation. It’s interesting that this is the first toulene/biphenyl protein to be found but it is also able to catalyze other similar materials whose pathways have been previously described.
I hate reading about how we as people are really destroying our planet. Antarctica has been brought up plenty of times in my previous Biology class about how bad it gets contaminated with oil and such.
I some what understand whats going on here. However, I am a little confused on what was suppose to be the outcome from doing this?
I am glad I am learning some of the vocabulary in our lecture class. At first I was reading this so confused, but then what a chemotaxis was and how it is increasing or decreasing concentration of a particular substance. Now I understand a little better now.
It is very interesting how they are able to use Phenanthrene or Diesel-Fuel and it become a sole carbon sources.
It is interesting how the D43FB failed to show the chemotactic responses, but still showed promising results.
I knew chemotaxis was important, but this paragraph alone shows how chemotaxis plays a significant role in experiments.
Why didn’t S. xenophagum D43FB not able to produce biosurfactants?
It is pretty cool that Antarctic soil holds such a range of bacterial diversities.
I think this paragraph is really interesting. Especially the recycling of hydrocarbons.
So SIP is basically used to label the DNA of organisms that use substrates?
I think it is interesting how they characterized the core.
I find it interesting how we did this in lab with the 16S rRNA.
SIP has been very interesting to learn about and to explore in this paper.
What are OTUs?
It is interesting that the six hydrocarbons and the two sediment samples yielded at a very low level of mineralization.
I like how this paper concluded, it is usually hard to understand some of the material and research being put forth. However, this paragraph helped me understand the overall point being made.
With the anoxygenic photosynthesis is this saying that light is still captured and turned into ATP but without the production of oxygen?
I find it very interesting how the bacteria are using the As(III) as their electron donor in place of water. Ive never heard of this before.
I believed I heard about purple photosynthetic bacteria before. They are Gram-negative I think, and make their own food.
I think RFLP is a technique that exploits variations in homologous DNA sequences that might have helped group them.
It is interesting how only a single clone type was detected.
Interesting that archaea were slightly more diverse.
Interesting to see that Ectothiorhodospira are dominating the Paoha Island biofilms.
Acetate is a removal of a proton correct?
Normal
0
false
false
false
EN-US
JA
X-NONE
/* Style Definitions */
table.MsoNormalTable
{mso-style-name:”Table Normal”;
mso-tstyle-rowband-size:0;
mso-tstyle-colband-size:0;
mso-style-noshow:yes;
mso-style-priority:99;
mso-style-parent:””;
mso-padding-alt:0in 5.4pt 0in 5.4pt;
mso-para-margin:0in;
mso-para-margin-bottom:.0001pt;
mso-pagination:widow-orphan;
font-size:12.0pt;
font-family:Cambria;
mso-ascii-font-family:Cambria;
mso-ascii-theme-font:minor-latin;
mso-hansi-font-family:Cambria;
mso-hansi-theme-font:minor-latin;}
I can see the obvious advantage bioremediation has over physicochemical treatment. Clearly any time we are able to avoid adding more toxic chemicals to the environment we should. However, I am curious about the disadvantages of bioremediation. Will the addition of these microorganisms disturb the balance in the ecosystem? Will other organisms be negatively affected due to increased competition for resources? What happens to the microorganisms after they have fulfilled their purpose of degrading the oil? Will they simply die off? Is this method costly?
Normal
0
false
false
false
EN-US
JA
X-NONE
/* Style Definitions */
table.MsoNormalTable
{mso-style-name:”Table Normal”;
mso-tstyle-rowband-size:0;
mso-tstyle-colband-size:0;
mso-style-noshow:yes;
mso-style-priority:99;
mso-style-parent:””;
mso-padding-alt:0in 5.4pt 0in 5.4pt;
mso-para-margin:0in;
mso-para-margin-bottom:.0001pt;
mso-pagination:widow-orphan;
font-size:12.0pt;
font-family:Cambria;
mso-ascii-font-family:Cambria;
mso-ascii-theme-font:minor-latin;
mso-hansi-font-family:Cambria;
mso-hansi-theme-font:minor-latin;}
If the oil entered and polluted this body of water nearly thirty years ago, surely it has spread and is affecting an even larger area by now. Is it even possible for bioremediation to make a significant impact on an area this large? How long would it take to see progress?
Why were the seawater samples transported on ice? What is the advantage of doing this? I would have thought that it would be best to maintain the samples’ original temperature in order to keep the bacteria alive.
What was the purpose of filtering the seawater and then adding the filter paper to the nutrient medium as well? Couldn’t you just skip the filtering step and have the same result?
The article says that eighteen strains had an adequate growth rate for further testing. However, they only sequenced the 16S rRNA gene for twelve of these. Why weren’t the other six sequenced as well?
If I understand correctly, BATH % indicates a strain’s ability to attach itself to the naphthalene; and it is this attachment that facilitates emulsification. I see that for most strains, their BATH and emulsification values are fairly similar, except for strain N4. Strain N4 has a BATH % of only 5.71, but its emulsification activity is still 62.23. What other methods could this strain be using to degrade naphthalene without direct attachment?
What further research needs to be carried out before this information can be applied to environmental cleanup techniques?
Is this gene (1, 2-dioxygenase gene) common to all naphthalene degraders?
Since the most important gene in PAH degradation is found in a plasmid, how common is it for non-PAH degrading strains to acquire this capability through horizontal gene transfer?
What characteristics make this strain a better model organism for bioremediation of PAH pollution than other strains?
Why would they construct a phylogenetic tree based on the C23O gene sequence if horizontal gene transfer is known to interfere with its phylogeny?
The Bushnell-Haas medium is considered a defined medium because its exact composition is known.
Also, I know that defined media which contain only a single source of carbon are called “simple” defined media. Is there a specific term for media that exclude carbon all together (such as BH)?
From what I understand, this experiment tells us which carbon source best supports the strain’s growth. This is determined by observing the turbidity of each microplate (which each contain different carbon sources). Higher turbidity means more cell growth. More cell growth means more degradation of that specific carbon source. Correct?
Why were E. coli strains cultivated (non-phenanthrene degraders)?
According to Table 1, strain ZX4 oxidized higher percentages of L-asparagine and D-glucosaminic acid than all of the other carbon sources.
Does phenanthrene become toxic to the bacteria at concentrations greater than those described here?
This paragraph states that strain ZX4 could be used as an engineering bacterium for producing indigo. Does this mean that these bacteria could produce indigo dye on a commercial scale and eliminate the need for extracting indigo from plants?
So is GST function basically similar to that of catalase, (which removes hydrogen peroxide wastes), except the GST removes phenanthrene components/wastes?
So does As(III) oxidation only occur during oxic conditions? And does As(V) reduction occur only during anoxic conditions?
So does arsenic oxidation and reduction occur best under anoxic conditions?
Why are the error bars for samples incubated with sulfide so large?
The specific line I want to focus on is “this results in a significant decrease in species richness and evenness, and a large decline in soil biodiversity of contaminated soils.”
Immediately after reading this, my mind shifted to think of a place we are all familiar with–the Mobile River Basin. The largest inland delta system within the United States is filled with an immense amount of species richness. I must say, many of us are not as familiar with Antarctica as we are with Mobile, Alabama. This led me to look into factors affecting our home town in hopes of relating it back to the study. Firstly, the Mobile Bay has a high contamination of water which leads to dead zones. These dead zones have such a high level of nutrients that the cyanobacteria becomes harmful to the region. When looking into Antarctica, the dead zones are claimed to be from the continuous burning of fossil fuels (which is also outlined in this paper). It is fascinating to see that these dead zones spread across all of the oceans across the globe.
The final thing I want to address in this paragraph is the source of these contaminates. Specifically, the addition of paper mills along the Mobile basin. This past year I had to opportunity to work for a paper chemical company directly in the paper mills. This personal connection intrigued further research. Many plants use a bacterial mixture in their waste water treatment before releasing back into the adjacent river. It turns out that this antimicrobial concoction may have a negative impact on animal and human populations. To the best of my knowledge, there are no paper mills in Antarctica, but it does not change the fact that mankind is the source behind the fossil fuel use in the region.
[by directly seeding contaminated sites with pollutant-degrading bacteria (bioaugmentation). Since the Antarctic Treaty impedes the introduction of foreign organisms into the Antarctic continent, bioaugmentation can only be implemented by the use of native microbes]
Since being in this class, I have solely focused on the idea of hydrocarbon degrading bacteria after being in lab. After doing some intense googling about my previous comment referring to dead zones, I happened to come across a nitrogen degrading bacteria. Many petroleums are able to be degraded using hydro-carbon degrading bacteria as listed in this paper. SAR11 is a nitrogen degrading bacteria that has been discovered to aid in denitrification. These bacteria consume nitrate and convert it into nitrite, in turn becoming gaseous nitrogen. This nitrogen then leaves the ocean releasing greenhouse gases into the atmosphere. The enzyme nitrogen reductase is a genome that potentially allows bacteria to breathe nitrate instead of oxygen. In fact, they have tested this on E. Coli and the DNA did produce the enzyme. Apparently, SAR11 is one of the most abundant bacterias in the world’s oceans. With that being said, there is a high possibility this may be a native microbe to Antarctica.
This is a very surface research level on the nitrogen degrading topic, but I am curious in how this compares to the carbon degrading bacteria. This would be a fascinating conversation to have with others to bounce ideas off of.
I thought about this particular oil spill as well. I thought it was interesting because everyone in the States heard about the disasters this entailed, but it wasn’t until a few days ago that I learned about the diesel oil problems in Antarctica. Specifically with bioaugmentation, the temperature of the waters in both regions are vastly different. Since typically microbes respond better to warmer climates, would there be a slightly different process based on where bioaugmentation is used?
[Samples were taken from the surface soil horizon (0–10 cm) from four sites exposed to diesel fuel]
What would happen if a deeper depth of the soil was analyzed? It states that they are taken between 0-10cm, but the severeness of diesel fuel exposure may be different at each sample site. At the different sites, is there a different maximum depth for alteration, perhaps maybe 15cm? I think it would be interesting to look at the results if samples were taken deeper than 10cm to see the effect of the naphthalene degrading bacteria.
Another thought on the soil depth–were the samples taken 0cm, 2,cm, 4cm, etc.? This seems like a way to see the difference in the saturation of diesel oil.
Polycyclic aromatic hydrocarbons are nonpolar compounds with delocalized electrons. My thought process is the idea of like attracts like. The water is polar causing the extremely polar oil to stand out. It seems that the bacteria may have to target nonpolar compounds. This is something I attempted to look up, but did not get a clear cut answer.
When doing some research for this topic, a new question arose. In colder waters, there is a higher amount of nutrients present due to the deep water rising to the surface, hence more phytoplankton present. Does the higher level on nutrients in the Antarctic water have a role in the non-polarity of the PAH?
The paper states “The optimal growth temperature was determined by a previously published protocol (Gallardo et al., 2014).” I looked up the study and it brought me to a writing called “Simultaneous effect of temperature and irradiance on growth and okadaic acid production from the marine dinoflagellate Prorocentrum belizeanum.” It states that this study was run at 18, 25, and 28 degrees Celsius. This agrees with the fact that the M&M we are reading was grown at 28 degrees Celsius.
[Bacterial growth in soil from non-contaminated sites was promoted when glucose was added, but not in control conditions or when phenanthrene was present]
When looking at the figures that are provided in the unexposed site, it can be seen that glucose did indeed encourage bacterial growth. It did start to plateau as the time (in hours) increased but I am assuming that is due to the decreasing nutrients presence as growth continued. This trend line is very apparent. In fact, right at the 72 hour mark, phenanthrene suddenly has a positive growth so much that the statistical bars do not over lap with the non-growing control group. When doing research on the metabolism of PAH, it was fascinating to see that this is how carcinogens initiate lung cancer. There must be an activation by xenobiotic-metabolizing enzymes for this carcinogen pathway to begin. The whole process of metabolizing PAHs is interesting when seeing the potential negative impacts.
It is interesting because Figure1D, shows that E43FB has the second highest phenanthrene degradation level. It has solid growth in the liquid M9 medium with phenanthrene but terrible growth with the diesel fuel (comparable to E.coli). However, the optimal growth temperature was determined to be 28 degrees Celsius. The average temperature in Antarctica is -49 degrees Celsius. E43FB had the most extreme cold temperature growth around the 4 degrees Celsius range. This leads me to your question: if I could predict, I wonder if we would see a change in the strains and they all flip growth rates since E43FB favors the cold temperature. It would be neat to even look at it at its ideal temperature and compare the growth there.
After looking in to xenobiotics too, it was interesting to see that these foreign materials would be extremely dangerous if it was not for metabolism. The PAH degrading bacteria helps to return the environment to the base condition. There are multiple enzymes that must be used in order for them to be properly processed. In fact, in the phase I biotranformation (in the CYPs) is where the metabolism of hydrocarbons takes place. The molecules present in phase II are generally less toxic and go through a conjugation reaction. Finally, there is a phase III but from my understanding the research is limited on this topic. It seems like this is the best place to explore and see the potential for PAHs and their enzyme-inducing properties. Since we can only have native substances in Antarctica, where are some other places we can pull enzymes from?
Also, when they say researchers pulled the most potent PAH metabolizers, does this mean they have a greater affect on larger hydrocarbons such as something like tetraphene?
Honestly, when I first started reading and found out we were looking at PAH degraders in Antartica it shocked me because I just never assumed anything grew in that region of the world. Knowing that archaea tend to be extremophiles I got to wondering if there was a higher population there over bacteria. I found out that 34% of the prokaryotic biomass was made up of archaea. This leads me to a question about arctic bacteria as well: in the harsh winter season, do some of these adapted bacteria become dormant? The weather can get down to -35 degrees Celsius and salinity of 37-327%.
Building off of this, I think looking at the endogenous and heterologous reporter genes is also very beneficial to the function of the bacteria. The housekeeping genes shows a lot about the metabolism under a variety of conditions. The heterologous reporter genes show the function of the promotor and how that can affect the bacteria.
Looking up upwellings mentions the process of circulating deep water/nutrients to the upper layer. I looked up the process of this in regard to nitrification. It looks like most of the studies are done on warm and oligotrophic open ocean waters. This studied mentioned comparing these upwellings off of the coast of Taiwan to typical coastal upwellings. When looking for the answer to your question, there was a mention about the turbulence of the water. It seems that growth of the microbes is more unfavorable in turbulent conditions.
I agree with this. It is interesting that in this study about upwellings that they are able to survey in the same location, yet the oxygen levels are able to vary. This will lead to less variables in the results than if multiple locations were surveyed.
I believe there is an increased nitrogen and phosphorous concentration in the Black Sea leading to eutrophication. This is due to increased fertilizer use in agriculture and improper wastewater treatment. Cyanobacteria are very prevalent in these waters.
[Total dissolved Mn were determined with inductively coupled plasma and optical emission spectroscopy]
It makes sense that heavy metals are being analyzed as they play an important role in inhibiting the nitrification process. Manganese oxide has a negative effect of nitrifying bacteria. If more of the heavy metal is present, we are most likely going to see a decrease in the amoA gene.
[Sequences retrieved in this study have been deposited in the GenBank under accession numbers EF414229–EF414283.]
I went ahead and went over to GenBank and typed in the accession numbers. I was interesting to look over this under the title of “Uncultured crenarchaeote clone BS100mB1 putative amoA protein-like (amoA) gene, partial sequence” for accession number EF414229.
This is really interesting because it is a natural phenomenon. I also saw that iron can come from the melting of icebergs. It seemed odd to me that iron, a metal, is found in the ocean itself. That is super neat that you talked about this in your geology class, specifically how the dust can travel such a far distance. That seems crucial as majority of the world is made up of the oceans. That reminds me of the Saharan Dust plumes that travels to the coast of my home is Florida. That makes complete sense that the iron comes with that, and I just always thought the sunsets were beautiful!
[the species that produces the siderophore with the highest affinity for iron can enjoy a competitive advantage ]
What happens if the siderophore captures too much iron for the bacteria? Looking at hemochromatosis:the human body absorbs too much of the iron. Is this possible for the bacteria when there is this competition due to the affinity?
I looked this up too. I also was curious what effect recombination had on antibiotics. It turns out that recombination plays a major factor on multi-drug antibiotic resistance. This makes sense as the genetic material is being shared through the genetic material. For chloramphenicol, it turns out the acetylation can prevent it from binding to the target (acetyltransferase).
[15 min for 24 h]
Just a quick comment, but the amount of time that has been involved with this study is crazy. Everything is done so precisely and there are many short times that must be tended to.
[Cell debris was removed from the lysates]
Being new to all of these research techniques, I wasn’t sure why lysing a cell would be beneficial after performing all of the experiments. It makes total sense when you look at it from wanting to maintain the highest yield. How are you able to control specifically what is lysed. After the centrifugation, can further separation be done from the supernatant?
[Geneious]
It’s really neat that there are these resources available now these days that allows for sequencing. This program was founded in 2003. It’s crazy to think about that a lot of these genetic discoveries are not even 100 years old. The discovery of DNA as genetic material was in 1928. The structure of DNA wasn’t even identified until 1953. In my biochemistry class we talk about the protein data bank a lot and it reminded me of that.
Continuing on with the next few sentences: I think the author brings up a good point that with the rich media that there is more than enough iron present within the media. There was exponential growth seen with the minimal marine media, but with the rich media I would predict a constant growth.
I agree with all of this from my analyzation. The increasing concentration of iron results in decreasing siderophore production. This remains consistent with figure 3B with the decreasing relative fluorescence as iron is added. The data also appears accurate as the error bars represent 3 replicates of the standard deviation.
I think it’s so interesting how species are so competitive that they evolve into such a strong species that outdrives the other. Not only is its own self getting stronger through iron uptake, but limits other species to such an extent they can no longer grow.
I was thinking about this too. Where is the limit that causes the bacteria to “switch”. And what if each bacterial cell has a different limit and some are producing while other are cheating.
[the energetic cost to WT V. fischeri ES114 of producing and using these components (Fig. S10A, B). The fact that V. fischeri maintains these functions, despite the cost, further supports the notion that they play a crucial role in survival.]
What are the odds that over time the cell develops a better way to conserve energy. It seems like the common topic in all of my biology courses is to make the most energy possible, but also conserve it. Perhaps an enzyme that could help with this function. I feel like this could lead to an even stronger competitive species.
The deeper the water is, the more less oxygenic. This means that the processes are not as readily available to occur. Therefore, the correlation is that the higher light transmission, the more oxygen is present (closer to the surface).
I think it is the presence of NO2, but also the presence of an oxygen molecules. In a, there is no oxygen present anywhere, but in b and c there is. It makes sense that when NH4 and NO2 are both present 14N15N continues in the upward trend.
Building off of this, it also is interesting to look at the graphs and see the narrow zone of net nitrification as mentioned. It shows that quick and sharp peak seen the strongest in the suboxic zone.
Here to agree with this comment as well. Think about the adaptations organisms have to have in the deep sea environments. Also think about how so much of the ocean is undiscovered. It is hard for us (humans) to explore the deep ocean, which makes sense why is hard for organisms to survive. For sure the nutrient level is a factor. We saw that in some of the previous figures (like why the suboxic zone is the best for net nitrification).
That’s so interesting! I had never heard of any of those techniques prior to this class. Bioremediation is particularly interesting to me. I think this is a field of research that can be incredibly beneficial. Bioremediation could be used to clean up oil spills or possibly target all of the fertilizer runoff into the delta.
I do not see us making much of a difference ecologically if we do not stop relying on oil as our #1 source of fuel for everything. In recent years there has been many efforts to “go green” and be more eco-friendly and while those changes definitely are making a difference, I think ultimately, if we want less contamination and prolonged resources, we need to re-evaluate our current methods. Good news is that research is always being conducted. Maybe someone is our class will figure out a solution for the oil crisis, who knows?
I think it would be really beneficial to collect samples from more than one fuel-contaminated site in order to get the most accurate data representation. In order to get a comprehensive data collection, this research should be conducted annually in order to monitor the abundance of soil bacteria with the growing oil industry. If these samples were collected on the 49th exploration, then studies should continue every exploration there after. The results would allow the researchers to see if the bioremediation that occurs would cause these harmful toxins to decrease over the years. The research conducted in Antarctica can then benefit the rest of the world.
R2A was the selected media for the analysis of this bacteria because of how well bacteria grows and differentiates on it. How would this experiment be hindered if a media was chosen that suppressed growth in some cases? Do you think there is a media that could have been more successful than R2A?
I think we would see less growth on MacConkey (selective of Gram -) and PEA (selective of Gram +). If those agars were chosen, we would be missing out on half of the growth. TSA agar though, I am not quite sure. When I performed a google search, it seems as though most researchers prefer R2A over TSA for a few reasons. While both +/- grow on TSA, there has been roughly a 16% reduction observed in the growth of colonies on TSA compared to R2A. It has also been thought that TSA can be contaminated more easily.
I found this very interesting, here is the link to the paper I read if you are curious: https://academic.oup.com/ndt/article/14/10/2433/1806023
I really liked your observation. This is great insight to know about “why” and “how” they conducted this study.
I find this very interesting. It was a bit hard for me to understand with how they worded it though. I couldn’t tell if they used the SSMS approach and it only isolated the most potent PAH metabolizers or if the researchers were just informing us of a new technique that could be used for later study. If this is a technique that was not used, I think this would be so beneficial to re-conduct the experiment using this substrate in order to get a full picture of all of the PAH metabolizers present in the Antarctic soil. While it definitely is great to understand the most potent of the PAH metabolizers, xenobiotics need to be studied more as a whole. With their impact on environmental health as well as human health, more growth in this field is needed to better understand their impact and how they influence the health of the organisms around them.
Hey Rachel,
From what I understand, in situ is different than continuous culturing. When in situ culturing takes place, the membranes allow for the exchange of growth factors and nutrients thus stimulating the bacteria’s natural environment in the culture. In situ conditions would not be steady, they would fluctuate like the bacteria’s natural habitat whereas continuous culturing is a way to maintain constant rates and conditions over an extended period of time.
I was curious as to what a chelator is, so after some research I found that it is a ligand(s) that bind ions/molecules to metal ions. It is often used as a therapy for lead poisoning, but other than that, chelation is widely debated. In this case, the siderophore in V. fischeri binds to the iron present in the water/media preventing V. harvey’s growth. This is a problem because iron is essential in order for V. harvey to grow.
I wanted to see how iron specifically inhibits the growth of these bacteria. Iron deficiency can lead to structual changes in many cells as well as decrease DNA/RNA synthesis. It is also essential is many metabolic processes and products. I found an interesting paper that walks through these processes in more detail if anyone is interested in reading it.
https://iubmb.onlinelibrary.wiley.com/doi/pdf/10.1016/0307-4412%2883%2990043-2
The use of both nutrient-limiting and nutrient-rich medias will allow the researchers to elucidate the necessary nutrients that are required in the natural environment in order for the growth and survival of V. fischeri and V. harveyi. This will be beneficial in understanding their development and allow them to monitor their growth under different environmental conditions (i.e. conditions that they flourish in and conditions that they dwindle in).
This is interesting! I just started researching this myself haha. That is definitely a neat way to combat tumor growth.
We use T-tests all the time in my directed studies lab. They are particularly useful to identify significant differences among the data and allows us to compare multiple features to see which pertains to the largest significant difference. Meaning we can compare multiple factors to see which one has the largest influence on the data.
This graph shows the growth of various genes under different settings. We see that in graph A, when 4 of the 5 genes are deleted, production of V. fischeri is greatly reduced, showing that they activate siderophores. To further prove that gene luxT, yebK, fre, and glpK were activators, when placed in the supernatant of V. fischeri is graph B, they grew exponentially because siderophores were present. In graphs C and D, we see that luxT reduces the translational reporter compared to the rest of the genes.
Graph A shows that something present in the V. fischeri culture inhibits the growth of the vibrio species. To begin understanding what exact inhibits their growth, the researchers test the growth of various bacterial species under V. fischeri supernatant. They observe that 3 of the 5 species were inhibited by something present in the V. fischeri. This illustrates the need for further testing.
It is very interesting that an aerE is present in both the cheater and non-cheater. Why do you think aerE would be present if you are not producing siderophores?
I do think you can, however this is definitely a part of the experiment that should be re-done a few times to observe the results. If the growth defect occurs multiple times, I would then say it is sufficient to deem it that possession of AerE makes an aerobactin cheater immune to cytoplasmic aerobactic toxicity.
Yes there is! Oxygen levels decrease with ocean depth, so there is a higher percentage of dissolved oxygen available at the surface. OMZs are found deep in the water column – typically around 700-1000 meters.
I too am very interested to see how this plays out in the rest of the study! I would hypothesize that the OMZs have just enough oxygen available for the aerobic ammonia oxidation to occur and begin the process of nitrification.
I am interested to see if the results reflect that this process does undergo steady state diffusion as the researchers just assumed this. If it is unsteady, that would make for a very interesting scenario for compound fluxes and when nitrification could occur.
Yes this makes sense! I wonder what their results will conclude and if there is a lower abundance of heavy metals in low oxygen/sulfide zones.
Figure 1D shows that nitrification by ammonox bacteria cells occurs within a narrow window of the water column – that window being suboxic conditions. Meaning that the concentrations of Sulfur and Oxygen are present in extremely low levels where nitrification takes place.
This phylogenetic tree displays the species of bacterial and crenarchaeal amoA found at different depths of the ocean. We see that majority of the species were found at 80 meters deep and decreased as we descended through the water column. The * also marks the species uniqueness in comparison to others in that zone. I suppose this is because of the characteristics needed to nitrify at that level under those conditions.
I wonder why we see more diversity within 80 meters as compared to deeper depths. I suppose it is because of nutrient availability. It’s also interesting that BS160B4 was found at 80, 100, and 110 meters!
Figure 1D shows that the anammox bacteria levels are much higher in the suboxic zone. This shows that nitrogen is consumed the most at this level as opposed to deeper depths.
“The degradation of PAH is usually initiated by hydroxylation, especially dioxygenation, which is catalyzed by oxygenase”
Hydroxylation is the 1st step in the process of breaking down PAH’s I was confused before about the difference and or how both process went to start the process. But I did some research outside of the article from https://www.tocris.com/pharmacology/oxygenases-and-oxidases and https://www.sciencedirect.com/topics/neuroscience/hydroxylation and I found that there are several enzymes responsible for the degradation 2 enzymes are a tick for tack situation and are both needed to complete the process.
What did they mean by Island that was kind of over my head did they mean the bacterium was found on an island or it like has a specialized niche in degrading phenathrene?
” bacterium Delftia sp. Cs1-4″
I understand that both pollutants are in the soil but I wasn’t entirely sure by their wording but are they basically saying that the PAHs are combining with the heavy metals to cause an entirely different substance or are they just saying that both pollutants together are the problem.
I guess another question is assuming that my 1st question was wrong and they aren’t combining with the PAHs even though the Heavy metals are present with the PAHs why are they unaffected by the pollutants of heavy metals?
Last thing I didn’t see them mention a simpler method they didn’t necessarily say what the PAHs needed to maintain their hold on the soil a simple solution to this problem could just be turning the soil exposing the pollutants to the atmosphere or changing the components of the soil which could cause the PAH to degrade through natural causes without microbial interference.
The article stated that “dioxygenase (PAH-RHD) and catechol-2,3-oxygenase (C23O) have been identified as the two key PAHs-degrading enzymes” because they know the problems that could possibly arise from the contamination from the heavy metals can’t they basically do the same thing we are doing with mega and blasting and find where the problem is arise and genetically modify the degrading bacteria to basically side step the contamination assuming of course time, money and that’s the point of interference from the heavy metals.
(in short jus trying to see if a different method can arise to bypass this problem)
Im not entirely sure what Liquid chromatography is what’s the difference between liquid chromatography and gas chromatography?
2.21 Is essentially what were doing now with our degraders currently. This procedure is I guess easier for me to follow and understand because it’s actually what were doing in lab now. Since its been a little while since 314 this gives me a nice refresher.
I’m just assuming because I’m not entirely sure myself but maybe they were trying to imitate the actual condition of rain and other elements in nature because I wanted to know the importance of the cycles myself.
After taking McCreddie’s biometry class understanding the stats of experiments are easier to understand I wish they would have spent a little more time explaining their work in the Statistical analysis portion of the materials and methods because in reality you can perform any stat to make your data to say what you want it to.
Would LB broth based agar be a possibility for us to use for our projects this semester for our posters since we will be using E.coli as well?
3.2 describes the effects CU(II) had on the degradation capabilities of Sphinobium PHE1 and it goes on to explain that in MSM everything was degraded but it depended on the concentrations the ideal concentration seemed to be less than 600 mg/L.
It does but more specifically I guess it’s saying that xyLE plays a role in the importance of how this gene enables it’s performance.
Rye Grass seems to be the answer in combating heavy metals but as I’ve seen in previous comments plants can take up or absorb metals from the soils, another experiment could be switching plants and trying to determine the variation in plant species.
I see that you have said that you said that we’ve seen 2 of the three paths of the biochemical attack in terms of degradation the mono-oxygenation at the C9 position discusses it’s affects on 9-Fluorenone but is this dealing with the isotope of F I’m not entirely sure.
So just going off the 1st sentence of the final paragraph “dibenzofuran-degrading Terrabacter sp. strain DBF63 can also oxidize fluorene, thanks to a cluster of plasmid-borne catabolic genes.”So are they stating that other PAH degrading bacteria don’t have this gene?
I kind of think that may be the case because even though we haven’t identified all the genes or organisms responsible for PAH degradation but just going off of what I know about bacteria I would assume so because I know there are other organisms from different families would live in the same niche doing the same thing.
Why not use a TOPO Kit? I remember for my MST we attempted a pGEM Easy vector kit but I didn’t receive desired results so I would assume that the TOPO kit is more reliable.
Why not use the 2 step RT-PCR kit isn’t that one considered more accurate, even though it may be possible to introduce more contamination I assume their sterilization and transfer techniques far exceed what we the class would have but they choose the 1 step instead. And I don’t think that contamination was a role player in their choice in terms of 1 or 2 step RT-PCR.
I feel strongly that because of this technique they were easily able to determine family characteristics on how your organism in question would act the phylogeny is basically a way to check and give you a baseline for things to test for.
I think that’s correct, I googled mutagenesis and found that most of the things online dealing with this process also dealt with PAHSs
I think that is correct. that sounds right to me
So in short should the takeaway from this section be that FlnA1&2 are the dioxygenase that initiates the attack on fluorene.
Fluorine and phenanthrene are the preferred substrates for this microorganism, And based off of their findings even though its not optimum they still can use other PAHs (naphthalene) so can in short I say that all PAH degrading bacteria have the capability of using all PAH’s as a carbon source. I say that because of after the talk a Monday gathered that in the right conditions mutations could allow for strains to pick up the capability of using different carbon sources as energy.
This indicates that this Gram negative bacteria has a entirely new pathway to degrade PAHs. “FlnA1-FlnA2 was shown to be capable of catalyzing monooxygenations and angular and lateral oxygenations of PAHs and heteroaromatics that are not oxidized by DFDO”
So in Short Carbofuran is just a pesticide? I wonder how they decided to observe the effects of this compound and NP degrading capabilities? isn’t this like adding another pollutant to aid in another? That’s how the 1st paragraph seems. The Only thing I could come up with as to why they would even think to use a substance like this to counteract another is carbofuran and it’s organisms don’t last as long as NP.
After continuing reading I understand my misconception with some of the previous statements made in the previous comment. the 1st sentence of the second paragraph set me straight.
Were they indicating that there was a possibility that some fungi have the ability of degrading carbofuran? They identified several strains of Carbofuran degrading bacteria I wonder why they decided to use Novosphingobium KN65.2?
I originally thought they used Novosphingobium sp KN65.2 as the experimental organism.
They grew the bacterium on TSB which is the same thing we were using to grow our NP degraders on. Actually a lot of this procedure is very similar to the approach we took to preform our mutagenesis.
I was a little confused about the importance of the gene they selected played in the mutagenesis process. ((tn5)
So the reason they performed the mutagenesis on tn5 is because that’s where the resistance on km was located, they didn’t clarify that the microorganism was susceptible to km
in short this mutagenesis was done to determine the location of the genes responsible for carbofuran degredation
With figure 1 it showed that the 2 were statistically identical.
the tree showed an association between our experimental bacterium to other sphinomonad species
so summarizing everyone’s comments this strain is basically a specialist that uses carbofuran as a food source with the capability to use other things as food sources
so in short this is just a visualization of how the mutants grow on carbofuran.
If the purple photosynthetic bacteria were most dominant, why was there more interest in the red spring colored biofilms the ones sampled?
Since PAH bioremediation can clean up pollutants completely or partially, what happens during a partial clean up? Is the area treated with additional clean up techniques to ensure complete removal of pollutants?
In regards to my last question, if the additional elimination technique is used, do they have to wait until the results of the first are shown or can they anticipate the use of a second technique based on the chemical structure of the pollutants in the area?
How was the decision made to utilize only 3 oil contaminated stations? Several samples from several different stations seems like they’d have move data. What systems was used to ensure the marine sediment was successfully not contaminated?
What criteria was used to decide whether the bacteria shows significant differences? What method was used to identify the bacteria strains?
Did they include both gram positive and grand negative or just have the gram positive?
With the research towards the eco friendly solutions, what steps are going to be made? Will they run the process the same way the first was does and what type of degrading bacteria will they add onto next?
The similarity in isolates in this paper compared t0 the random choices of isolates in the last paper seems like the results would be easier to interpret and move forward with. Knowing that certain circumstances were used when choosing colonies would make it easier to decide whether the study went well because of certain phenotypic and genotypic qualities of the bacteria.
Since the colonies chosen were colonies that looked similar, did they find that when the DNA was sequenced, it showed similar DNA sequences?
Since it is a versatile bacteria, is it possible for it to every work against degrading PAHs and do something different? How interesting would it be if it was able to reverse the work that had already been put in place!
Finding metapathway genes in this bacterial species would be a great new study!
Do these bacteria work best degrading aromatic compounds or does it seem like it is best at almost any compounds since typically aromatics are more complex?
Are the bacteria derived from the hot springs manipulated to perform more closely to archaea? I’m assuming the extreme conditions would favor the growth of a modified bacteria that can withstand the conditions.
What caused the inability to obtain PCR products?
did they choose the different months to see growth during different weather patterns?
So would this suggest that the bacteria prefers reactions in the light? Seems like its able to degrade better in the light instead of the dark
Why as the rate lower after 200 hours of incubation
What techniques were utilized to clone the microbes and were all cloned or just the ones suspected to be top degrading bacteria?
How does C23O sequence the gene, is it a normal DNA sequencing program?
Is it the lack of salt in the Bushnell-Haas minimal salt medium that causes the soil samples to become agitated? What significance does this provide to the study??
Were all of the strains identified by similarity to a specific strain? That seems like it would cause errors because the alignments weren’t 100%, at least they weren’t stated to be.
What criteria was used when choosing whether it was similar? In other research based classes there was a certain percentage used as the minimum to choose the similarity.
What additional ideas were there to explain why there was no detection of arsenide oxidase genes?
Since there was no detection of the gene, what is the assumption for the cause of oxidation of arsenate?
Is it assumed that acetate favors light reactions
Would biological methods for treatment of pollutants introduce organisms that could potentially damage the ecosystem? Are the microorganisms selected for specific traits, such as nutrient requirements, from what microorganisms are already present in the affected environment or are they introduced?
Potentially, how would bioremediation occur with a microorganism suited for a highly specific environment? Would genetic alterations be possible to expand the organisms ability to perform set functions or is there a method to synthetically replicate the degradative processes for natural remediation of the pollutants?
What is the ratio of oil stations to area surveyed? Are the islands close with similar environments? Would a more locationally diverse, environmentally similar sampling system give better feedback for the bacteria present?
What is filtered out of the sample with the use of the 0.2 um paper filter? Just microorganisms larger that 0.2 um?
What is the rate of Napthalene degradation after 15 days? In regard to the experiment, does the degradation continue until all resources are gone without excretion or breakdown that could be harmful to the environment?
What exactly does the bath percentage specify? Also N16, N17, N18 all showed low growth (N16<N18<N17 0.201) with respect other cultures. Yet the emulsification activity varied between N16 and N18 such that N16 was higher than N18. Is it possible that a bacteria could be ill-fitted for growth in the environment but has a higher emulsifying capacity?
How would a nonromantic compound function in the same situation? Would it be degraded more easily the less stable the compound is?
Can certain strains of the naphthalene-degrading bacteria only degrade naphthalene that have certain genes (e.g 1, 2-dioxygenase gene)?
Can the conversion of catechols be catalyzed by a pathway other than the tricarboxylic acid cycle? And are the compounds involved in the cycle more or less effective than others?
What do the determination of mol% G+C and Biolog-GN test for?
Bushnell-Haas (BH) minimal salt media is utilized in the examination of fuels for microbial contamination and microbial hydrocarbon deterioration. The media excludes a Carbon-source but contains all other nutrients needed for the selected bacterium.
Within the medium, trace elements include Magnesium sulfate, calcium chloride, and ferric chloride. Ammonium nitrate provides a main nitrogen source. Buffers include mono potassium phosphate and potassium phosphate.
The BH medium can be used at varying pH levels, temperature, and salt concentrations to select for the strains with the highest degradation of hydrocarbon oil.
The liquid Luria-Bertani (LB) medium is a widely used media for the culturing of bacteria. I assume this would be used to grow pure cultures of the bacterial strains with the highest degradation levels in the BH cultures.
Phenanthrene is a polycyclic aromatic hydrocarbon (PAH). It is described as colorless, monoclinic crystal, with a faint odor. Occasionally, they appear yellow. Solutions often exhibit a blue fluorescence. PAHs are formed when oil products (coal, oil, gas, and garbage) are burned but the burning process is not completed.
Bacterial glutathione transferases (GSTs) are enzymes known for their ability to aid in cellular detoxification. They also play roles in protection against chemical and oxidative stress. Due to the chemical composition, they have the ability to catalyze nucleophilic attacks on the electrophilic groups of many hydrophobic, often toxic, compounds.
The C230 sequence is known for its ability in bacterial strains for degrading aromatic pollutants. It is considered an extradiol diogyenase, which incorporates oxygen into one substrate.
The focus on one highly effective PAH degrading strain is unusual in comparison to the last paper where the results of many strains were compared. Is it possible that this study was not as thorough in the broad view of potential PAH degrading strains?
The paper focuses on the relations of the ZX4 strain to S. paucimobilis which is relatively abundant in many environments. Would the study have began with research into the potential heightened PAH degrading qualities of these strains or a selection of strains and then research?
Putative promoter sequences mean the region is believed to function as a promoter but has not yet been proven to be the promoter. What relevance does this have to PAH degraders? Does the proving of a promoter sequence give more insight to how the strain is regulated?
How would the use of the meta-pathway operon aid in improvement of PAHs degradation capability? Could you use gene editing? Otherwise what type of gene modification is being referred to?
A chemoheterotroph is an organism that cannot synthesize intermediates of nutrients and therefore ingests inorganic substances to derive energy from. These types of organisms derive energy from the oxidation of their electron donors, which is why sulfide or H2 are essential.
Is shifting between the two states energetically favorable or energetically expensive?
An anoxic environment is one that has limited or no oxygen available. Therefore anaerobic respiration is most likely to be used to satisfy the microorganisms energy requirements. Mixotrophic organisms should be able to utilize anaerobic respiration and photosynthesis as a means to derive energy. Would phagotrophy or parasitism of any sort be found as well in this type of environment?
As the suspensions bubbled with N2, which is often the end product of nitrogen fixation, can we assume nitrate reductase is active in some or all of these organisms?
The arsenate respiratory reductase genes aids in the conversion of arsenic from a solid form into a usable soluble form. The cycling of arsenic can be influenced by arsenate (As(V)).
Would the amount of arsenate transporter genes increase or decrease based on a greater presence of As(II) or As(V)?
Are the lower rates of As(III) oxidation in the presence of light and higher rates of As (V) reduction in the absence of light indicative of the greater energy yielding processes? In light the As(III) oxidation which yields the highest amount of energy most energetically favorable in the given conditions?
Depending on the genes active can we identify and/or active certain regions to promote certain respiratory arsenate reductases?
Was there a test done in an environment that was strongly oxidizing or weakly reducing? Could that have a significant change persistence of the cyclic nature of the data?
So does this mean that sulfide or hydrogen are able to accept electrons to allow for the reduction of As(V)?
what is the CTAB method?
Is naphthalene more or less easily degradable than phenanthrene?
What makes the stain a good model organism?
Why does it need to be swabbed from the slant medium?
What does Sall do to the DNA?
Why couldn’t you obtain authentic PCR products for arsenite oxidase genes
Why does it shift over the 2 states?
why did you wrap them in aluminum foil?
Why did they take the green-colored sample up if they aren’t going to use it?
Why are the archaea more diverse?
how are As(v) and As(III) able to shift so easily?
Why are dark As(V) reduction a broader range of temperature than the Light-driven As(III)?
This paragraph discussed the little process of the removal of crude oil from aliphatic components. What exactly is the method of bioremediation? Also, what makes PAHs resistant to biodegradation?
As discussed in this paragraph, the Nakheel beach region did report PAH biodegradation that was not isolated nor investigated. Present times results that there is no evidence of the bacteria strain in that specific region. What could have been the cause of that?
What exactly is the Kimura 2-parameter method? I understand that it is use for a phylogentic tree to establish evolutionary distances. But what is done to contain those results?
PAH degradation I am assuming was used in order to determine the bacterial growth on the NA plates. I understood the other chemicals, but what exactly does inoculum solution do?
Considering that LC grew “poorly” on pyrene and phenanthrene and LB grew on all hydrocarbons except pyrene. Does that make LB is more useful for hydrocarbon bioremediation than LC? Since that it mentions that strains that grew on PAH substrates can be useful for hydrocarbon bioremediation.
Rod bacteria are better than coccoid bacteria because how the rod bacteria contacts with the surface and utilize settled or dense particles. Rod bacteria also divide faster than coccoid bacteria. Is there a way to tell if the population of rod bacteria is greater than or less than the coccoid bacteria?
Interesting to read that the accumulation of HMW can increase the cell weight to where it settles at the bottom of the liquid making it less turbid. What is the purpose of growth assessments?
From looking at the table 1 location A contains less essential nutrients which decreases the productivity. So would location B by looking at table 1 would have the most productivity due to being nutrient rich.
Within the vent fluids are recyclable hydrocarbons? What exactly is the petrochemicals and how is it a suitable natural model system besides that is migrate upward to the sediment surface?
Confuse, so in this studied it talked about studying PAH-degrading bacteria in sediment cores so then how is the hydrocarbons being identified within that?
When the paper reference core, what exactly is the core?
For colonies forming a clear zones, I assume are the most suitable for the experiment. What exactly does it mean for colonies to form a clear zone, is there PAHs present?
What I got from the introduction paragraph is that selenium when in found in the ocean can be very toxic when there is high levels and can cause severe poisonings to the fish.
The objective for this is experiment is to identify gene that can reduce Se. In past studies Enterobacter cloacae SLD1a-1 is known to reduce Se(VI) and Se(IV).
All the strains were placed on an LB agar, did all of the plates contain the antibiotics? When it says that the “antibiotics were added supplements to the medium” is it meaning extra nutrition for the strains?
The rate of reduction of selenate was calculated by observing its concentration medium. Aliquots of the culture was used in ion chromatography to be analyzed and filtered.
The LB agar is able to identify clones that can perform reduction on Se(VI). White colonies are identified as clones that are unable to reduce, while bright red colonies are identified as clones that are able to reduce Se(VI). In the experiment the results showed that there was no change in color medium with E. coli S17-1, does that mean that the clone did not react, but why was their no white colonies?
If the fnr gene was unable to transform Se(VI) to Se(O) why was ogt gene not used in this experiment to see if it would transform Se(VI)?
FNR is a transcription regulator that is active when oxygen is not present. It makes physiological changes in the cytoplasm. What is the importance of FNR in this experiment
In conclusion of this discussion, that the precipitation of Se(O) can be changed in “”suboxic conditions” by oxygen-sensing proteins. FNR is a potentially control for Selenate reductase genes.
How long would it take the proper bacteria to completely clean the contaminated environment?
Would added the degrading bacteria to polluted environment affect other microorganisms that also live in that environment?
Would this procedure be similar to Sergei Winogradsky enrichment culture technique? Reason I asked this is because they used a selective filter naphthalene to eliminate autotrophic bacteria.
I feel like this is an important part of the research. It could also shield some light on my question I asked in the introduction “What would be the amount of time needed for the bacteria to completely clean the polluted area?” . I think the concentration would have a great effect of the rate it takes to clean the area. Hopefully there will be more about it in the results section.
I have great reason to believe there is a positive correlation between percentage of naphthalene degradation and emulsification activity.
Based on figure 4 600 ppm or high would be considered harmful to the selected strain because growth at these concentrations are even lower than 2oo ppm which is a fairly low concentration.
High cell surface hydrophobicity and production of more emulsifier allows the N1 and N7 stain to be more functional in higher concentration.
Would the gram positive cell wall of the bacteria allow it to be more acceptable in the highly variable environment of oil-contaminated tropical intertidal marine sediment.
As established, 400 ppm is the optimum concentration for napthalene degradation for both gram positive and gram negative. Because gram negative cell wall is more complex would the growth rate of the gram negative bacteria be higher?
Based on the study provided I strongly believe gram negative bacteria would be a great help in cleaning contaminated sites compared to gram positive bacteria.
Would PAHs have a negative affect on the human body?
The degrading mechanism in marine bacteria may be slightly altered from the terrestrial bacteria of the same spices.
This could be a great way to compare the differences between the marine and terrestrial bacteria.
What was the genomic library used for?
I feel that this is an important step in characterizing the isolate.
Transduction is a common method used to transfer new DNA into the organism studied.
I am curious why they want this gene to be expressed by the lac promoter?
Could this gene be found in other similar species?
Having the enzyme dioxygenases allows it to be aerobic.
Because this specific gene is found in different location in different strain, it shows that this characteristic is optional for survival. The reason I believe this it because in one strain the gene is found in the plasmid which carry non essential information and in other strain the gene is found in the chromosome.
When looking at how the isolate amino acid sequence of phnA1 fell out of the major cluster you must take into consideration the environment.
Because phnA was not able to convert anthracene and monocyclic aromatic hydrocarbons, that does not have a big effect on its ability to transform other aromatic hydrocarbon such as naphthalene and 1-methylnaphthalene.
How does the chemoheterotrophy use these inorganic substances differently than the photoautotrophy? Does it have an effect on growth of each type?
What other ways could they have examined the cycling of arsenic under anoxic conditions
The processes of nitrification and denitrification is based on the same principle as the coupled reaction of As(III) and As(V).
This is an idea location to find archaea being that this is such an extreme environment for other microbes.
All PCR’s were preformed in triplicate to insure the needed DNA was amplified. This is a good technique when analyzing DNA.
When looking at the temperature range and optima for anaerobic As(V) and As(III) other factors should be considered not only light and dark and temperature.
From fig 1 I gathered in light As(III) decrease as As(V) increase in concentration. In a dark environment As(III) increase as As(V) decrease in concentration.
Are Ectothiorhodospira sp. commonly found in shallow well oxygenated environments with extreme temperature?
I agree with their detection of oxygen-dependent As(III) oxidation because the sample was collected in a shallow area which is prone to have a high source of oxygen.
Since temperature is a main factor and greatly affect As(III) oxidation and As(V) reduction how does the light and dark contribute to the overall performance of As(III) and As(V)?
Are you suggesting that a colony of bacteria be resistant to different environments all at once? For example, a colony of bacteria could be resistant to an environment with high salinity and high temperatures. If this is possible and the resistant DNA is known for these extreme environment factors, maybe plasmids could be used to splice the DNA into the genomes of the bacteria. However, if the DNA isn’t useful to the bacterial genome, it could later be spliced out.
Maybe multiple simulations were run to test for any potential environmental effects. Also, the products of PAH bioremediation are already present in the environment. They likely won’t cause any new issues.
Is it possible that the different nutrients available at the depths the samples were collected could from could influence genetic mutations in the bacteria collected?
How difficult is it to reanimate the stock bacteria? Do most of them survive?
Because the bacteria were cultured in plates with naphthalene as the only carbon source, any bacteria should have been naphthalene-degrading bacteria. Could the bacterial colonies found to have insufficient growth on the initial plate been more successful when cultured on a plate of a different level of naphthalene?
Based on Fig 4, all strains showed maximum growth at 400 ppm, but growth began decreasing for all strains after 400 ppm.
Growth rate and percentage of naphthalene degradation do not appear to be directly correlated at 200 ppm. Strain N7 has a higher growth rate than strain N1, but strain N1 has a higher percentage of naphthalene degradation than strain N7.
It’s most likely that the bacteria they’ve identified can utilize other carbon sources as well. While they can survive using naphthalene as their sole carbon source, it isn’t the only option.
Your hypothesis seems plausible. We had to force our bacteria to degrade naphthalene in lab by reducing or removing other carbon sources. N7 may more readily use naphthalene as a carbon source, especially when naphthalene is the only carbon source available.
It would also be intriguing to see the similarities between the naphthalene degrading bacteria collected in the Persian Gulf and those that could be collected from the Gulf of Mexico.
Based on the text, the claim that they are only partially studied means the bacterium haven’t been extensively researched. Based on the research that was available at the time, they were unable to determine whether marine bacteria or terrestrial bacterial were more efficient naphthalene degraders. I believe this is a way of sort of narrowing the research topic.
How do the products these degraders produce affect their environment if the PAHs degraded were carcinogenic, genotoxic, or cytotoxic?
DNA is filtered across the agar by size. Smaller pieces travel faster, so the bands visible near the end of the gel farthest from the wells is short sequences of DNA. Bands near the top of the gel are longer sequences.
A low concentrated agarose gel is necessary in this experiment because the DNA sequences are fairly long. This allows for the long but still shortest sequences to travel farther down the gel. A higher concentration of the agarose gel would have likely resulted in wide bands near the top of the gel.
A holoenzyme is expressed if it can breakdown substrates into products. If E. coli is able to do this after the gene has been inserted. The gene should already encode holoenzyme. Genes inserted into a genome in this experiment shouldn’t be mutated.
Transforming the substrates was the test to determine if E. coli had incorporated the holoenzyme into its genome. It also showed how extensively the holoenzymes can breakdown substrates.
Plasmids are nonessential segments of genetic material. While they can often offer the cell antibiotic resistance, bacteria may not necessarily maintain plasmids if the genes are nonessential. Bacteria only need antibiotic resistance when exposed to antibiotics.
The cell likely consumes less energy maintaining multipurpose electrophoresis proteins. Fewer proteins would need to be synthesized, and there would be fewer pathways for mutations.
Based on my understanding, PhnA1 does not code for the small subunit it needs for full monooxygenation activity. Perhaps if combined with PhnA2, it would have a useable small subunit for monooxygenation activity.
The presence of multiple dioxygenase genes, I believe, is what allows the bacteria to degrade a range of aromatic hydrocarbons.
Are high concentrations of arsenic toxic to the uncontaminated soils in Japan? Is a particular enzyme needed for the reaction being studied?
There are a number of other amplification techniques that could potentially be useful alternatives to PCR such as multiple displacement amplification, loop mediated nucleic acid sequence based amplification, isothermal amplification, or strand displacement amplification.
The biofilms are photosynthetic. They are likely stored in the dark to prevent photosynthesis in the new environment away from arsenic.
A clone library is a type of DNA library. Complementary DNA fragments are cloned and inserted in host cells to create the library.
The sample of arsenic being reduced is a radioactive isotope.
The DNA was sequenced and compared to sequences stored in BLAST. The results should be fairly conclusive.
What would be the results of an experiment using varying levels of both As (V) and As (III)? The figures depicted only seem to show either As(V) or As(III) as being present and the other being absent.
The breaking of a hydrogen bond leads to higher energy production which is why the Figure 4 shows As(V) being reduced more quickly in the presence of add H2 than in the presence of sulfide. The reduction potential of H2 is more negative.
Water in more shallow areas is constantly being moved. Like in low tide, areas that are usually covered in water are exposed to more oxygen. In deeper waters, the soil or anything growing near the bottom isn’t exposed to much oxygen because the water doesn’t move in the same way.
I interpreted it more as redox rates decreased as toxic byproducts built up, killing some cells. I didn’t think they were ever really measuring bacterial growth rates.
yes, that would make it easier for the PCR to be ran.
Why is there less information on higher molecular weight PAHs? what does the molecular weight of the PAHS have to do with the degradation? has it never been done or how does it work?
Is the Persian Gulf the only place this can be done? it seems as if it more pollution being done. have they tried anything other than the bioremediation? has it been done effectively to insure more help than harm?
If different zones of the Persian Gulf are polluted why do they keep making it a port for oil. if it had been polluted since the war why keep polluting the gulf? I think more pollution test should be done and I think the bacteria need to be researched due to the fact they may be causing problems as well.
why are the samples taken from different depths? do the researchers know exactly what depths to take the samples from to get certain results?
does the ONR7’s various concentrations of naphthalene affect the bacteria growth? does it actually help give results or since it contains naphthalene does it just enhance the bacteria growth of the samples?
why was the supernatant put into the vessel containing anhydrous sodium sulfate? what does it do to the supernatant?
are the concentrations of naphthalene added to ONR7 to enhance the bacteria curve? whydo they add different amounts?
why do the aromatic compounds with more rings get decomposed faster? what do the rings actually help in the aromatic compounds if the more you have they are decomposed faster?
how did the gram positive bacteria aid in the degrading of naphthalene in the oil- contaminated topical sediment? does the cell wall allow the bacteria to act accordingly in different environmental settings?
do the harsh/different environments cause the il degrading bacteria? is it created as things take place in different environments? why is it always separated?
what was the hold up in actually observing and testing the bacteria until now? is the naphthalene common?
what is a environment with only gram negative bacteria? s it a livable area? what do you think caused it to be all negative?
I was wondering the exact same thing. what makes it so special that it is only gram negative?
are naphthalene degraders not eco friendly? what could help with future oil spills that is eco friendly?
since the PAHs are cytotoxic, genotoxic, and, carcinogenic to the marine life, once humans consume the seafood are the PAHs none existent after cooking or are they slowly doing harm to the human body? How can consuming raw marine life or killing marine life that has been affected with the PAHS affect humans?
How do the scientist know when they are placing the specific genus in an area that the area will have the specific carbon sources required to get the degrading done properly?
Indole crystals are very common I see we even use them in lab! but using the crystals do they differentiate the indigo coloring and Naphthalene degraders? and with E.coli not being used as a PAH degrader is this really a smart move?
what is a cosmid clone? why are clones and sub-clones being used in the experiment? will the clones have mostly the same results as the original or does this make the results vary? the lac operon will tell the results of their orient?
If a high concentration of agarose gel would have been used do you think the reults would have been recognizable ? although they would be at the top of the band in a wide sequence do you think it would have been able to be purified?
why do you think the order of the phnD genes changed? does it have something to do with the environment or chemicals around?
I also have that question Cheykola! I was wonder if it threw off the results in any way but it seems maybe they drifted from another but still formed together.
why was it necessary for them to be hydroxylated just to be degraded? does it make the degradation an easier process?
I do not think the gene is needed. I think that naphthalene degrading bacteria can do the process on its own if need be.
They found small sub units, just not in their regular location the chart 3 says they were found in phnA2.
what is Sau3AI used for in this experiment?
Why do you think the genes encoding the PAH dioxygenase were located in a separate transcriptional unit? are they more advanced or hazardous?
Due to the genes having multiple dioxygenase is this good r bad? do we want them to only have one? what harm can be done to the genes or experiment if genes keep being found with multiple alpha subunits?
so do you think if the scientist put down unfit primers in multiple areas, it will hinder the results? do you think they actually start over and put new primers there?
I googled what a NanoDrop spectrophotometer was and I did not know it assessed the purity of DNA as well as quantified it. it allows the samples to be pipetted.
Are clone libraries the same as DNA libraries? DNA libraries are DNA fragments that have been cloned into vectors so that researchers can identify and isolate the fragments.
is the only difference between radioassay incubation and regular incubation the antigen being measured in radioassay incubation?
why do you think the dominant clone group was closest to the 16S? Do you think the reason it possessed the ability to grow anoxygenic photosynthetic oxidation of arsenite from the biofilm but the other clone group was not?
It is weird that the sample could shift whenever the light and dark shifted. how did the sample know when to adjust or when it reverted back the first time why did it keep switching instead f sticking with the last shift it stayed with? is it a special trait the sample possesses that allows it to sense the change?
Sha’Torrie,
I looked up this question, in 2010 Felisa Wolfe-Simone led a team through a similar experiment
If the type of oxidation was ancient, do you think other scientist before found linkage and didn’t understand?
I looked up what the Cre-lox method was and it is very interesting. It is a site specific recombinase technology. It is used to carry out deletions, insertions, translocations, and inversions in specific DNA sites.
Does it matter or make a huge difference that P73T was grown in artificial seawater? Will this create a difference in sequencing or analysis of the strains?
Isn’t the 216L agar plate a blood agar plate? If so would it be better to use a nutrient agar plate since they are better for growing bacteria?
protocatechuic is a dihydroxybenzoic acid.
this would be a great attribute to the experiment so the different bacteria can be seen.
why do you think they said it would be interesting to study the role of 5 plasmids? do you think this would help further experiment?
why would the functional genes be used as indicators? is this more beneficial to the process?
PHE is a polycyclic aromatic hydrocarbon composed of 3 fused benzene rings
what is abiotic stress? How would it affect the process?
Thank you! that was a great definition.
Maybe the heating or cooling of the cultures at the wrong temperature. UV Radiation, or even exposure to air could affect these cultures.
An HPLC is a High Performance liquid chromatography technique. This is used to separate, quantify, and identify each component of the mixture. It also relies on the pumps to pass a pressurized liquid solvent containing the sample mixture through a column.
If the soil contained no PHAs shouldn’t this be a benefit to the pot experiment?
Qpcr are very interesting due to them being in real time. this is a beneficial method to the quantification process.
This project was much easier to understand. The experimenters seem to know what’s going on and have less variables which makes it less difficult to keep up with.
C230 was amplified out of the strain this shows it has a higher concentration than the other bacteria in the strain.
why is the degradation of PHE dependent upon enzymatic activity? Which is more beneficial more or less movement?
So do you think iron or nickel is actually more beneficial in the degradation process? I’ve always heard iron but not nickel.
This has been the clearest and least confusing discussion paper intro. I understand exactly what they are about to do.
we already know from the previous paper this Sphingomonad strain will yield results and assist with PAH degradation.
should the experimenters fear fluorene being attacked again due to the attacker enzymes not being identified?
this paper came at a great time because we have began discussing electrophoresis in lab.
why was the plasmid cloned so many times? what were the benefits?
I think they dissolved the extracts to keep them sterile.
when I researched this topic it says the name it was under was furadan and it was banned due to the toxicity of this insecticide. it seems no one is using it in cropping fields.
Carbofuran has a very long half life but it is degraded by hydrolysis, if the soil is organic it will speed up hydrolysis which will cause a quicker degradation process.
This is interesting and gives an amazing outlook on the current random mutagenesis experiment.
why were RNA extracts diluted prior to the conversion?
when it comes to degrading anything the best thing to do is find the best pathways. using liquid chromatography should help because it separates proteins, nucleic acids, or small molecules.
did they only describe the first version because it was the better option?
an inducible enzyme is only under conditions in which it is clear of adaptive value and constitutive enzymes are produced at all times.
the pathway probably had to be constitutive due to carbofuran being so prevalent in majority of the pathway.
how are roles assigned to gene functions?
were the amplicons created naturally or artificially? does that make a difference in the results?
Photosynthetic organisms are photoautotrophs, which mean that they are able to synthesize food directly from carbon dioxide and water using energy from light
What other bacteria does strain P73T contain?
Why are metabolic pathways so rare in marine environments and projects ?
The use of the BLAST system to build phylogenetic trees seems to be common. Actually using this technology before allows me to understand why it was used in the experiment and how it helps with protein analysis.
What is reverse phase bonded silica? I’ve never heard of this in my biology courses ?
So was the Sphingomonas strain used as a comparison ? As in was it used to compare strains found? Did they use the similarities in other strains to decide they were degraders ?
Why were so many different fatty acids found in 1 strain?
Horizontal gene transfer is the movement of bacteria from unicellular to multicellular, why do you think the bacteria switched back and forth between the two?
How is P73 able to degrade compounds such as PAH?
What are truncated reading frames?
Why did it encode the lower pathway ? Was it set up that way or did it just happen ?
What is meant when they say genetic aquisitions are the cause of it being able to catabolize?
I had to research gene knockout because I had never herd the term. Gene knockout is a genetic technique in which ones of an organism’s genes is made operative.
The last step they did with further cloning, is that what we will do with Marcus’s clones? Will they then be sent off for further sequencing as well?
isn’t self formed adaptor PCR used for chromosome walking? why was it used here?
Yes, iron does help PAH degradation. its a type of Anaerobic degradation of PAH.
I know Im slightly late to the discussion, but upon reading through only the introduction and the comments I noticed a lot of people mentioning the BP oil spill in relation to this topic and couldn’t help but think, “Wow we must be responsible for a significant portion of that microbial destruction in Antartica and probably all over the world!” In addition to the oil spills (we know of) the United states is in fact the largest producer and consumer of oil, consuming and producing almost 20 million barrels per day according to the Energy Information Administration of the U.S. (eia.gov). That is an absolute insane amount and the need to shift from fossils fuels and protect the remaining microbial diversity in Antartica and all over the world is an issue that deserves first priority. Just really makes me think how much destruction we have already done that we cannot undo.
I love this commentary Jessica!! In addition to using the proposed solution of using the natural abilities of bacteria, I found it very interesting that although scientists are restricted from introducing foreign organisms to Antartica’s soil under the Antartica treaty, using the native microbes is actually the “best method of choice in the bioremediation of soils with low indigenous PAH-degrading bacteria.” So I also feel it is as you said, “the earth is trying to fix itself/self-heal”and unfortunately we may need to catalyze the process.
This is not something I had not taken into account before when considering the method of PCR. It sounds very logical since DNA has a double stranded helix that is antiparallel so a forward and a reverse primer would be necessary to copy the 16s gene on the corresponding strand. I did a little more research out of personal interest and it turns out The 27F primer would anneal to the antisense of the double-stranded helix (3′ to 5′ direction) and the 1492R primer would appeal to the sense strand of the double-helix (5′ to 3′ direction).
“Polymerase Chain Reaction (PCR).” Diamantina Institute, The University of Queensland, 9 Feb. 2018,
Aliqouts from the four diesel fuel-exposed soils and the four non-exposed control soils were eventually isolated and incubated for 72 hours at 8, 18, and 28°C. Would the Phenanthrene degrading bacteria from the collected sample cultivate more efficiently while incubated in conditions more similar to the environment of King George Island at lower temperatures or would growth be ideal for the bacteria at higher temperatures?
“Bacterial growth in soil from non-contaminated sights were promoted when glucose was added, however not in controlled conditions or when phenanthrene was present. In contrast phenanthrene supplementation of a diesel-exposed soil promotes sudden bacterial growth after 72h, suggesting that phenanthrene is being metabolized and used as a carbon source.”
This excerpt made me wonder how some microbes adapted or were forced to develop in diesel contaminated soils to have the ability to degrade the polycyclic aromatic hydrocarbons . Were these microbes already present in the soil, if so why is growth not promoted by bacteria in the non-contaminated soil using phenanthrene (even if it is a scarcely detectable amount)?
Reading a little further ahead it seems that the polycyclic aromatic hydrcoarbon degrading microbes were present in both samples/soils all along (since 111 were isolated that originated from non-diesel exposed sites out of 350), suggesting that the (PAH) degrading microorganisms flourished over a period of time in the contaminated sites and allowed for exceptional strains of phenanthrene-metabolizing bacteria to develop. The three highest strains from the contaminated soil isolated have the ability to degrade Phenanthrene 69%, 86% and 95% This is a spectacular amount considering the dangerous carcinogenic effect of the (PAH).
I agree with you Jordan. I think the process of microbial bioremediation is the key concept that can be carried over to alternate situations for other endangered ecosystems. The three degrading bacteria genera isolated and observed to be the most efficient degraders of PAHs in this study are restricted to only native bacteria, so there is no telling how many different bacteria with the same ability to degrade PAHs remain undiscovered.
“Several PAH degrading strains of the Sphingomonadaceae family have been isolated from soils from the southern end of the Argentinian Patagonia, ratifying the presence of this type of PAH degrading bacteria in cold environment soils.”
This make me question wether some PAH degrading bacteria are able to live in warmer climates or if any have been discovered. Is the cold environment essential to the process of PAH degradation?
Autumn, I am also unfamiliar with the term “marine oil snow” and thought this excerpt from the University of Delaware did a nice job of explaining this natural phenomenon.
“If you were able to stand on the bottom of the seafloor and look up, you would see flakes of falling organic material and biological debris cascading down the water column like snowflakes in a phenomenon known as marine snow.
Recent disasters like the Deepwater Horizon oil spill in the Gulf of Mexico, however, have added a new element to this natural process: oil.”
Marine snow can include fecal matter, sand, and other inorganic particles in addition to dead animal and plant material. I think that in this case the oil is somehow attaching to these particles, as more accumulates the density increases, and the oil seep down at a faster rate which introduces large quantities of potentially damaging oil to the seafloor.
The deep-sea has received considerably less attention in respect to reports on hydrocarbon biodegradation since the first published reports in 1970. This statement implies to me that there was such an exponential increase in the demand, production, and damage accumulated from oil that we had to dynamically expand research on hydrocarbon-degrading bacteria to keep up with the absolute horrid destruction accumulated from hydrocarbon pollution. Especially after the Deep Water Horizon blowout! Although I think it is beautiful to watch the resolve of the scientific community (such as the proposal/research of bioremediation), it is upsetting that this research stands in our current circumstance as further exploration and production of potentially dangerous hydrocarbons in deeper water continues.
Was 21°C chosen as the second incubation temperature because it is more similar to the average surface water temperature? Or possibly for maximal bacterial growth in the lab?
This was a little difficult to understand for me as well. The internal standard makes sense, but I am unsure about how they would proceed to differentiate between them. u
I suspect it was quite a surprise when the measurements reveled that ”all six of theses PAHs were mineralized by the 4567-24 sediment sample” conversely to the oil rich core 4571-2 which was expected to have yielded higher mineralization levels than that the quite oxidized 4567-24 sediment core.
Ok, so looking at the graph helps to make a little more sense as to how the endpoint selected for extraction of DNA from 13C incubations was 11 days. I can clearly see a progressive removal of PHE occurring around day 9 followed by an asmptote around day 11.
Shelby, this is a great way to set up look at possible future experiments involving these two samples. I would think that the bacterial community from sample 4571-2 would probably be more efficient in mineralization of PAHs in a laboratory environment more similar to its own because of the exposure of oily substances found in the surrounding environment. I think this would definitely be worth pursuing.
I want to make sure I understand this correctly. So essentially of the 68 sequences, 3 OTUs were identified, and OTU-1 comprised the majority of the 68 sequences, so the genus was determined based on the identification of the gene for PHE degradation? Secondly, was this reinforced by the abundance of 16s rRNA genes for SIP clone PHE1 (OTU-1) increasing by magnitudes during incubation?
Wow they already basically remodeled the process of coupled arsenic metabolism under aerobic conditions with an already defined coculture “consisting of aerobic As(III) oxidizer strain (OL1) and an anaerobic As(V) respirer (strain Y5).” I think thats pretty significant and it seems they want to focus this investigation on the phenomena under only anoxic conditions.
It seems Arsenic may be added to this list of biogeochemical cycles of elements as it seems to display the capacity to partake in these coupled reactions that give energy for growth and are essential for some life! I think it is fascinating these microbes can resume a “normal” existence with a metabolic activity that is so alien and it only makes me wonder what else may persist on our planet that could possibly withstand the extremes of other planets.
I was thinking the same thing! “Red-colored biofilms dominated by purple photosynthetic bacteria”? Not only that but it seems that these red biofilms were actually found more common which is quite interesting.
Since the arsenate respiratory genes (arrA) were PCR amplified I am assuming this gene is found in both the archaea and the bacteria since representative clones were sequenced. I was wondering if these genes would be located in similar loci.
Correct me if I am wrong, but in this figure it displays the cooccurrence of Aerobic As (III) oxidation and As(V) reduction both in the presence and absence of light, however the oxidation of As(III) in the presence of light is visibly slower.
As (III) oxidation can be achieved via anoxygenic photosynthesis so I think the fluctuations of these concentrations are a result of the energy source being influenced.
This introduction paragraph explains the topic well and the current research into PAHs. However, it does not clearly state the research that has been done here. The previous paragraph tells us what we need to be looking for in an good introduction, but thus introduction fails to answer the question: What did they do?
I noticed that the paper has pointed out multiple times that there is not as much known about PAH degradation in aquatic environments.
Are we supposed to comment on this section as well? Do you want us to read the background information and comment on that as an assignment?
I believe that is part of what was explained in class Friday. In order to determine the genes responsible for PAH degradation, we need to be working with a fully sequenced genome. So in this study, where they have isolated samples from various oceans, they must first determine the specimen’s genome before they can determine PAH degradation and genes responsible.
I know that it says that more information is available on all aspects of the mutant generation method, but I wish more detail was given as to how they decided which gene to isolate for deletion. If we do not currently know which genes are responsible for PAH degradation and we are searching for more answers to locating the genes, how did they decide to delete this one in the P73T strain? I would assume more trial and error at several attempts altering genes in other strains before a successful attempt. I would not expect a detailed explanation in this section, but mentioning briefly how it was selected would help to answer, if I plan on conducting a similar experiment, where do I begin to determine which gene to isolate?
Even with your tip, it is difficult to derive too much information from these figures. I understand that the circular maps indicate that there is a GC skew because of the 6th ring in the figure, but are we to deduce any information from the magnification of 60X?
Does the fact that the P73 strain is composed mostly of unsaturated fatty acids also aid in its uptake of aromatic hydrocarbon compound uptake? I would imagine if the membrane is more fluid, then what genes are responsible would be able to control the fluidity of the membrane such that it could allow for uptake of larger hydrophobic substrates as needed.
I wonder if this has to do with the majority of the research involving non aquatic bacteria. If the P73 strain had needed mobility to reach PAHs, it would have had the genetic material to become mobile. However, if the environment were rich enough to produce enough without having to move, it would be biologically wasteful for it to devote so much energy. If the marine environment was indeed rich enough and the current provided all the mobility our bacteria needed for uptake, it could have regressed the mobility adaptation.
I’m not really sure what I am supposed to take away from this image. Please explain.
Why is it only productive along that route? What happens if 1-hydro-1,1a-dihydroxy-9-fluorenone is not formed? What about the pathway leads to this formation over another?
Thought I would share the results of my wikipedia search for some background for my peers in case they needed some further information on sphingomonads.
Sphingomonas was defined in 1990 as a group of Gram-negative, rod-shaped, chemoheterotrophic, strictly aerobic bacteria. They possess ubiquinone 10 as their major respiratory quinone, contain glycosphingolipids (GSLs) instead of lipopolysaccharide (LPS) in their cell envelopes, and typically produce yellow-pigmented colonies.
The sphingomonads are widely distributed in nature, having been isolated from many different land and water habitats, as well as from plant root systems, clinical specimens, and other sources; this is due to their ability to survive in low concentrations of nutrients, as well as to metabolize a wide variety of carbon sources. Numerous strains have been isolated from environments contaminated with toxic compounds, where they display the ability to use the contaminants as nutrients.
Due to their biodegradative and biosynthetic capabilities, sphingomonads have been used for a wide range of biotechnological applications, from bioremediation of environmental contaminants to production of extracellular polymers such as sphingans (e.g., gellan, welan, and rhamsan) used extensively in the food and other industries.[5] The shorter carbohydrate moiety of GSL compared to that of LPS results in the cell surface being more hydrophobic than that of other Gram-negative bacteria, probably accounting for both Sphingomonas’ sensitivity to hydrophobic antibiotics and its ability to degrade hydrophobic polycyclic aromatic hydrocarbons.
So both gram-positive and gram-negative bacteria are capable of PAH degradation, but fewer data is available on gram-negative bacteria. The reason that there is less information on gram-negative bacteria is because they were until recently believed to be unable to use PAHs as their sole source of carbon?? Is this correct?
Thus, the significance of this study would be to prove gram-negative bacteria capable of oxidizing fluorene and open a greater area of study in PAH degradation.
I was not sure what BamHi, NotI, or NsiI enzymes were so I was reading about them. They are restriction enzymes cable of recognizing short DNA sequences and cleaving them at the target site.
The supernatant and the pellet were remixed together or they were separated then mixed separately?
What is the significance of the presence of pyruvate?
Should be take any special note to the bit about fluorene degradation by protocatechuate in the lower pathway? None of the other information in this paragraph seemed to flow with this bit of information, yet it seemed to stand out to me.
When doing a quick lookup of what protocatechute is the product of protocatechuic acid and O2. Protocatechuic acid is great for pulling off O2 (looking at the structure, it would readily undergo hydrolysis), but the protocatechute did not have much information.
I know that you spoke briefly in class about the insolubility, but I am not sure that I completely followed. I understand heat’s affect on enzymatic activity and that IPTG triggers transcription, but how is it that it goes from being insoluble to soluble?
Does a gram negative bacteria taking on genes from a gram positive bacteria take on more traits from gram-positive bacteria like outer cell membrane? And would that change its stain pick up when trying to stain a sample to determine the cell type?
What is DFDO vs CARDO?
The table also shows that ability for these pathways to produce alternate products. Do these products need to be taken into consideration when choosing ideal PAH source?
How was the group of genes determined?
This introduction is very brief. It seems to end abruptly.
Is the fructose here used as a supplemental carbon source like the glucose or pyruvate in previous experimenst?
EASYTRAP is a system of recovery of DNA from agarose gel using the principle that glass powder absorbs DNA and allows for simpler and quicker collection of target DNA fragments. It’s more than 50% efficient and uses lower melting temperature.
This makes sense. It paper 2 the LB126 strain was found to be closely related to other Sphingomonas species and Pseudomonas.
This seems like a very inefficient method of determining gene expression.
consensus motif is the most frequent order of residues (either nucleotide or amino acid) found at each position in a sequence alignment.
Why is the mononuclear Fe2+ binding domain important?
What does it mean it is realted to another large subunit of PAH dioxygenases but it did not fall in deep branch with other PAH dioxygenases? Either they have left something out here or I am getting hung up on this wording.
Does this mean that dioxin dioxygenase, ferredoxin, and ferredoxin reductase are all required or only required to be coexpressed in the E.Coli strain?
The arhA1 gene was inactivated. If the mutant is not involved in PAH degradation, all that is saying is that it doesn’t know the gene’s other cellular functions? I would assume if they knew arhA1 was inactivated, they would no longer be interested in it for PAH purposes.
Is there a way to look at a a comparison between all (or a fraction) of known PAH degraders, their location in the genome. and what PAH they degrade all in one location to be able to visually examine possible connections?
Do all Sphingomonads retain these listed characteristics even if they are the recipient of genetic material from horizontal gene transfer? Or could the obtained genetic material change these “typical” characteristics?
I believe this is the first time we have seen it discussed that there has been an isolation of transcriptional regulators. Would this have been done using RT-PCR to detect expression then genes isolated based on the findings of the expression (in a previous experiment)?
I don’t know that it would be necessary to sequence the entire genome. I think that since the genes involved with the first phase of degradation have already been isolated, they could examine sections of the genome that work in unison with those genes by looking at expression. Creating clones with various sections of DNA which also include the genes involved in the first degradation phase, then plating the clones and examining the colonies for PAH capability, sections of DNA containing the 2nd phase of PAH degradation can be selected for sequencing. This method would be less time consuming and less costly than sequencing an entire genome. Plus I would image it would be difficult to locate the genes encoding the electron-transport of that particular protein even looking at the entire sequence.
Why have they chosen a filter-mating method? I remember reading about this in genetics, but I’m not sure why you would choose this method of conjugation over any other genetic transfer.
Unable to view T1. Clicking this takes you to a link which asks if you want to download this paper.
Why were they transcribed in the same direction?
Can we break down the logic behind the ligations into the various plasmids? Why did they have to ligate these genes (arhA1A2, arA3, arhA4) into pBBad22T, pBBadA12, pBBadA13, and pBBadA14 plasmids on;ly tp introduce all the plasmids into A4-PCM1? Is it much like the concept of separating the arhA1A2 and ferredoxin/ ferredoxin reductase?
How would these results be if all 4 mutants did not produce a clear zone?
In reference to Table 3:
It says that ORF17 is part of ORF11 (as its source) but the %identity shows homology from 2 different micro-organisms for ORF17 and ORF11…
What region are they posing was deleted to create ORF11?
So there is no evidence in the database where the C-terminal region of ORF6 originated?
If it does not have the NAD-binding domain, is this ORF essential in acenaphthene degradation?
The 2.4-6.2 increase of mRNA arhA3 and arhA1 levels in the presence of acenaphthene, is this a good result? It seems rather low.
If the P73 gene responsible for PAH degradation was acquired through lateral gene transfer, but the B30 strain did not produce PAH metabolic capabilities, does that imply that P73 acquired that gene through HGT after the 2 strains evolved into different species and now continues to pass the gene onto daughter cells?
Why can they not use a radical reaction or ozone to break the c=c double bond? In organic chemistry, that was the easiest way to break double bonds even with an aromatic. Why can we not throw some O3/Br2/Cl2/ excited electrons in there?
Is it important that they are the only bacteria within one phylogeny family? This is just one family of proteobacteria. Are there other bacteria families capable of similar fluoranthene metabolism?
So we don’t currently know if the plasmids play any role in PAH degradation?
I honestly would have never known that pollution and oil spills can affect microbial communities. I usually will only think about oil spill and pollution by the beaches and warm oceans. Now that I’m reading just from this paragraph, I can see how oil spills can soak in the soil and destroy small microbiological organisms or destroy fossils. fossils
I am a little confused about the PAH of diesel fuel. Is the different levels of PAH diesel fuel is like the different acidic/ base levels? Is it considered the same chart range we all learned in chemistry? For example, in sentence three phenanthrene is the smallest PAH for the Bay region. This sounds like a PH level of acidic or base level.
Before looking at the comments, I was wondering why does it take so long to analyze the soil too. I was thinking that maybe it takes a while to see bacteria growth from the soil and after it finally grows, they have to separate it to find single colonies. And thinking of that scenario, the scientist have to do that for every base. However, now reading our professor comment, the theory of the length of the expedition makes way more since.
I honestly think it is amazing how these scientist take there time to analyze the culture samples. It surprises me how many steps methods was used to be able to see the colonies in the sample. Hopefully we can do and learn all this in lab just not in a three month time period.
As I am reading towards the end of the paragraph, it says that the scientists found over 350 colonies from the soil count. I just remember from lab 2 where if it was above 250 there is too many to count. I am jus wondering did they count the colonies by hand or was there a computer system that can detect how many colonies? Is tat was it meant by screened, like being ran under a machine?
In this paragraph it says that D32AFA and D43FB had similar growth pattern and used diesel fuel for a energy source. The E43FB did not use diesel fuel for an energy source. My question is what the E43FB use for a source or did it not need a source?
It says in this paragraph that D32AFA and D43FB had the similar growth pattern and diesel fuel was used as a energy source on that site. On E43FB it says that it was unable to use diesel as a energy source. My questions how did the workers use something without diesel fuel or did they use nothing at all?
Now that the scientist have found the three phenanthrene degrading strains, could they find these strains in other cold countries like Canada or cold year round cities like New York? I am asking this due to the sentence that says oil spillage is in cold weather sites. Oil is everywhere?
In this paragraph is it suggesting that scientist who have an interest in this research experiment, do they think that the UBiome company may have used there tested made up samples and not the official samples from the Antarctica sites?
I honestly thought that the oil from the oil spill either disintegrated, got burned off from the hot ocean floor, or cleaned up from the oil rig machines. I didn’t know after all this time oil is still adding to the sea floor.
My question is how do scientist study or collect the hydrocarbon degrading communities without getting burned, the machines getting over heated, or the organisms being burned during that process?
My question is in Figure one, why is it that the sea floor with the oil spill of hydrothermal cores seem rocky or rougher that the regular background core which looks smooth? Does the regular background core have oil spill present also?
I also recognize these primers and using PCR reactions. However, in lab we only did 4 PCR reactions. Its amazing how they are using 96 clones meaning each clone need to be tested with a PCR reaction. That is a lot.
I am wondering why couldn’t authors isolate the organisms from the mid Atlantic Ridge? Was the depth of the sea floor to low to be able to collect a pure organism sample?
I am having a hard time understanding the difference of heavy or light DNA? Does this depend on which bacteria was founded/
As from reading this paragraph, it seems that As(V) and As(III) are dangerous. I wonder what component or chemical nutrient that makes this element poisonous.
I also believe there can be high levels of arsenic in soil all over the world. Especially in countries or states were there are more oil spills or industrial work fields. There can be different levels in a smaller region also. This is what I assume.
I am confused on how does the scientist know the artificial medium will be exactly like the Pahoa Island hot spring water? Do they take a sample from the spring water and compare the chemicals to the medium?
I was wondering since the clones should be a copy of the 16S rRNA gene, how after the blast sequence the clones did not have similarities to the gene database? Is it by the 16S rRNA mutated, and therefore the clone was different than the original gene?
I was thinking the same thing. I was thinking with using the BLAST system in this experiment, every archaea will be slightly the same but, I guess not. Therefore, since there were four different archaea groups did the BLAST tell how did they become different in theory?
I am wondering could they change to another more efficient primer for a better application result so I can be a aoxB?
I think that is cool too. I wonder even through time and the evolutionary change of the earth will cyclic nature of the phenomenon still be the same or alter in some way?
Since the primers was not suited for the experiment, I wonder was the novel mechanism an unknown primer for the As(III0 oxidation?
Is there a bigger gap in knowledge about marine-sourced degraders because they are more difficult to study?
It is very interesting that they are able to isolate a bacteria from a deep-sea sediment! I am curious as to what made them choose a sample from the Indian Ocean or if this was just random!
This is very interesting to me! I think it is really cool that they are able to grow P73T in the artificial sea water and add in the flouranthene to observe and research it.
As a marine biology major, I am very interested in anything that has to do with the ocean. I have never previously put a lot of thought into bacteria in the deep-sea. I think it would be really cool if we were able to grow bacteria from sediment in the ocean in class! This has made me more interested in the bacterial growth, DNA extraction, and genome sequencing!
I am having a hard time understanding this figure. Maybe because this is the first time I have seen a figure like this. I understand the large one is the chromosome and it explains that from the outside inward is the way the rings are counted. Therefore, is it showing that the very first outer colorful ring and the purple and green ring (1&4) are the forward and reverse non-coding RNA? This figure is just very confusing to me.
What gave them the knowledge that the P73_0346 was the gene out of this gene cluster that needed to be deleted?
This figure shows again the evidence that the chromosome and plasmids have different evolutionary history but in a bar graph form instead of the phylogenetic tree. I like that they used a few different figures in order to show this difference in their evolutionary history.
I have seen several phylogenetic tress throughout my biology classes containing animals/organisms, but this is the first time I have seen one used in comparing gene sequences and proteins. I would have never thought to use this to show data when carrying out an experiment similar to this, but it is a very good way to show this information.
The fact that the P73 bacterium strain could be an aid in any future marine oil spills is very interesting. If this is true, discovering this PAH-degrading bacterium is even more rewarding.
This paragraph states that additional experiments are necessary and more exploration is needed to find more complex information on strain P73. What are these other experiments that will need to be completed?
It is very sad that this contamination is an issue from industrialization and urbanization. The fact that the coexistence of the PAHs and heavy metals can cause damage to the ecosystem and human health should make people want to reduce this industrialization and urbanization. Although most of the human population does not care to do something about this, it is reassuring that it is possible to use the bioaugmentation method to degrade these pollutants without causing other damages and needing too much funds.
I googled what exactly ryegrass is, and it is a very pretty grass. It also said that the ryegrass grows best in mid-winter, therefore, while undergoing this experiment this is a factor they must consider when planting the ryegrass. They must keep it between 5-18 degrees celsius when growing and testing on the ryegrass.
How were they able to determine that the soil didn’t contain PAHs or copper before collecting it?
This part of the experiment seems to be the most time consuming taking 84 days just to get the soil they needed to extract the DNA from. Is there any way of speeding this process up? Are the 4 wet-dry cycles over a 4 week time period necessary?
So they created this phylogenetic tree using the same process and program that we used to create our phylogenetic tree in class on January 30th, correct?
These results show that xyLE gene and the C23O gene are more commonly present with a high concentration of PAH, but when the PAH has been degraded those genes disappear. Is this showing that these two genes are involved in the PAH degradation process?
What does the direction of the arrows indicate? I know it says the arrows represent ORFs but it didn’t say anything about the direction of them. I noticed that the arrows for each structure are each going the same direction, but some are just more stretched out or have longer lines between the arrows. This is a figure I haven’t seen before so I wasn’t sure what I was looking at.
In this figure, it shows that the treatment with the PHE-1 and the treatment with the PHE-1+Cu are much lower in PHE concentration than the control. The treatment with just the copper is about the same concentration level as the control which shows the Sphingobium is definitely keeping the PHE concentration down.
In this figure, the expression maintains or becomes level in contrast to figure 2 in which we see the expression of the gene decrease. It is hard to completely compare these figure due to the large difference in the scaling though.
It is very interesting to me that they were able to find out this bacterial strain can help reduce the heavy metal and PAHs co-existing in soils. Although I am curious, is it sure to help with any plant growth like it did with the ryegrass?
Since the genes are dispersed throughout the genome, that makes this species more difficult to study than others. So in the previous two papers we read, the species they studied were not as difficult to determine the PAH degrading genes compared to this one?
The last sentence of this paragraph allows me to understand what they are going to be experimenting in this paper. I was partially confused when reading the first part of this paragraph so I am glad they stated exactly what it is they are going to be looking at.
What are the antibiotics that the strain is grown in overnight for?
Thank you, I understand this more now! This is also what we discussed today that we have to find the antibiotic that our bacterium is resistant to in order to complete our random mutagenesis experiment right?
The MEGA software seems to be a very popular software used for alignment and phylogenetic tree construction! This is the software we used and that the past 2 papers we have read use.
Is a truncated transposase a “shortened” transposase enzyme? As in some of the DNA sequences in this gene were deleted? I tried to google this but wanted to make sure I am understanding this term correctly.
In the last sentence it states that the strain LB126 may have been inherited by lateral transfer, this is the same as horizontal gene transfer right?
Their gel electrophoresis was not a very clear picture. I know they aren’t always perfect, but I think they could’ve got a better picture of their results than the one that is in figure 3.
In the last sentence of this paragraph, they state that the strain LB126 may have been inherited by lateral transfer, this is the same as horizontal gene transfer right?
So for our project, would we need to grow our bacteria strain on a PAH like flourene and on glucose to compare as they did in this experiment?
Table 2 is a little overwhelming. It has a lot of information in one table so it is kind of difficult to understand.
So for both Sphingomonas and Novosphingobium the catabolic pathway is unclear, but they choose to focus only on the Novosphingobium bacteria in strain in this paper.
This is scary how dangerous the carbofuran pesticide is to humans and other mammals! I googled to see if there were any deaths associated with this pesticide and I read that in Nigeria in 2004 there was one death but several cases of vomiting from the carbofuran residue on batches of noodles.
They state that triplicate cultures were incubated, does this mean they did 3 replicates of each culture? As in 3 replicates of the carbofuran-induced cultures and 3 replicates of the -noninduced?
They grew a culture overnight on agar plates by placing the paper filter onto the agar plates? Then they took the paper filter off the plates and placed into 25 mL 0.01 M MgSO4. I wanted to make sure I am understanding this part of the procedure because I have never performed or seen the filter paper on agar plate techniques.
So the pink to light red color showed up more on medium with just carbon as a source? Or did it still have both nitrogen and carbon but just less nitrogen?
So although this is saying it is closely related to the two different Novosphingobium isolates, that does not mean it has genes that came from those isolates by horizontal gene transfer? Just that it has genes similar to those that are in these isolates?
I can gather from these graphs that as the different mutants are added, the concentration of the formation of carbofuran phenol increases compared to graph a which as no formation. I am not sure what the take home message of these graphs is supposed to be though.
The sentence that states ” The insertions in mutants 6C1 and 21D7 mapped in the same contig.” Is this saying that these mutants did the same thing when inserted?
They decided to test the Novosphingobium to see if it grew on methylamine because the knowledge they had about the first step of the degradation of carbofuran being the release of the methylamine in a different bacteria, correct?
So the last sentence is stating that only the aromatic ring carbon in the carbofuran compound was mineralized as we talked about in class?
Is there a way to test their observation and if it is linked or not to the expression of carbofuran mineralization and of the cfdABCDEFGH operon?
This paper was very difficult for me to understand. So they did find the catabloic genes that aid in the degradation of carbofuran, but they could not identify the genes that are required to initiate the mineralization process?
What physicochemical treatment regimes do biological methods have an edge over? Are in situ biodegradation processes the best way to alleviate pollution even though they can completely destroy organic compounds?
Naphthalene is harmful but yet is found in a lot of common things. If Napthalene is so harmful, why is it used so frequently?
How would results change if the sediment samples were taken from about 50 cm below the surface and seawater samples were not transported to ice but were at RT?
What is the CTAB method used for DNA extraction? What else can the CTAB method be used for?
Why did most of the bacteria have no motility? What would have been the result if the bacteria were oxidase-negative?
If N16 and N18 exhibited the lowest levels of naphthalene at 200 ppm, what would have been the result if 20 ppm or 400 ppm of naphthalene were used?
How exactly is the bioemulsifier production screen test performed?
So, one of the main questions with this research is whether gram-negative or whether gram-positive bacteria plays the important role in naphthalene degradation?
Continuing the discussion from paper 1, I know that PAHs are pollutants generated during incomplete combustion of organic materials such as coal and petrol. Bio-remediation is a safe and effective way to eliminate these environmental pollutants by using microorganisms to consume and break down these pollutants.
It would be interesting to see what the phylogenetic trees for this strain looks like and compare to the phylogenetic trees of isolates obtained from the Persian Gulf in paper 1.
In this paper, the Biolog-GN plate was used, but in an article I read, there is another plate that is very similar to this one. The Biolog-ECO plate has a similar use to the GN plate, so why was the GN plate used over the ECO plate for this research? In the article that compared the plates’ ability to distinguish among microbial communities, it didn’t clearly state what the major difference between the plates was. It did state that the two plates demonstrated an equal capacity to discrminate among the micobial communities? Does it have to do with the nutrient media or is there another reason the GN plate was better?
From my understanding, the BH Agar contains all nutrients except the carbon source for bacteria growth. Each isolate was tested in BH liquid medium containing phenanthrene as the sole carbon source, but for the first 7 d, was the phenanthrene the sole carbon source as well?
Would the strain have grown on the MacConkey Agar since it is selective for gram-negative?
If LPS is essential to the outer membrane of gram-negatvive bacteria, how does this affect the outer membrane of S. paucimobilis if it lacks LPS?
Can xenobiotic compounds be metabolized via the salicylate pathway?
Why can S. paucimobolis degrade xenobitic compounds and not PAHs?
Nitrification is the conversion of ammonia to nitrate aerobically and denitrification is nitrates being converted to nitrogen gas under anoxic conditions.
What unique characteristic about periphyton made researchers think it would be the right complex mixture for arsenic detection? Was it the components or the function or both?
Why were green-colored springs and small ponds relatively rare in comparison to the red springs which were abundant?
Why was HPLC and spectrophotometry used to for the analytical determinations?Would HPLC not work in the determination of Sulfide?
As (III), which is the electron donor goes down when there is light and that means that electrons are being donated to the photosynthesis apparatus.
There is no electron donor in Figure 4 in the top picture. Since the samples that were amended with sulfide decreases, we know that there is sulfur oxidizing bacteria in the biofilm. The samples that were incubated in H2 used arsenate really quickly. The bottom picture shows the consumption of arsenate and regeneration of arsenite.
If chemoheterotrophy did not drive As(V) reduction, then what did?
Can Hydrogen and sulfide serve as electron donors for other growth of diverse arsenate-respiring prokaryotes other than chemoautotrophic growth like chemolithoautotrophic growth that uses C02 as its sole source of carbon for growth?
I do not like one bit the idea of ubiquitous, toxic pollutants tainting the environment which we live. And with potential routes of exposure including via the respiratory pathways, or dermal absorption the issue of these PAHs should no doubt be addressed. I am curious, how are these compounds prevailing so in the environment? Are they being introduced on a huge scale by human activity, or are PAHs forming more rapidly somehow because of an increased level of carbon in our atmosphere?
The genus Rhodococcus sound like bacterial allies in our efforts to remove some of the harmful contaminants maliciously probing about the environment. This key factor of possessing hydrocarbon dioxygenases, detectable in microorganisms by oxidation reactions is fascinating. Such eloquent use of chemistry, and now we have the ability to widely detect and isolate these dioxygenases. I wonder, is there a harmful by-product of PAH degradation?
What makes you think that they are looking for multiple genes?
“However, the genes for the intrinsic electron-transport protein for ArhA1A2, the genes for the degradation pathway of their metabolic products, and their regulatory genes, have not been isolated.”
Should this not be the true purpose of this paper? To isolate these genes?
It may not be relevant, but is the CFMM used only in these scenarios when we have to find an organism’s dependence on another chemical for survival?
More importantly, why is cDNA used when we use genomic DNA in our own experiment?
The table shows 90% amino acid similarity to Erythrobacter litoralis HTCC2594. How is the highest identity shared to Burkholderia sp DNT?
[By contrast, significant indigo formation was observed only for strain A4-PCM1(pBBadA14) (data not shown). These observations showed that ArhA3 and ArhA4 function as the electron-transport system for ArhA1A2.]
This shows that A4-PCM1 produces indigo from indole, to answer the question you asked.
I personally believe that random mutagenesis is a very important tool that can show us function of a very particular protein structure and it’s function and it could be useful to identify our genes in question.
I think redA2 is just a ferredoxin reductase gene that relates to Sphingomonas wittichii strain RW1
Yep, I believe so. The structural similarity of all of the PAHs means that their pathways are very similar.
I mean its a part of a whole new family, probably similar to other Alphaproteobacteria since Rhodobacteraceae are a part of that class.
I think that making mutants is the most straightforward procedure to begin with. Expression cloning and RT-PCR is complicated and requires known primers to conduct the procedure.
Metabolites can be tested by mass spectrometry like they do with urine tests in medical labs.
Kanamycin resistance genes?
What are you referring to Hampton? It does seem like they identified PAH degrading genes, based on paragraph 37 alone.
[Therefore, strain P73T, the first fluoranthene-degrading Rhodobacteraceae bacterium reported, may be a useful strain in which the C-7,8 dioxygenation pathway involving extradiol cleavage of 7,8-dihydroxyfluoranthene can be studied]
What exactly can this be useful for? Is the dioxygenation pathway particularly important for particular kinds of PAH degradation?
[Notably, region B of the genome, which contained the PAH-degrading genes and were absent in another bacterium of Celeribacter, strain B30, was predicted to have been acquired via lateral gene transfer.]
Maybe I need to look over my genetics and organic chemistry (possibly) but what is the region B in the genome?
To extend upon Shelby’s point about ways bioremediation can be used. Have studies been used to see if these natural microbes can be used in medicine? Has there been any attempts to see how they can decontaminate foreign/mutated cells to a human immune system?
I understand that bacteria and other minerals grow in soil, which is easily found and extracted in warmer climates Though, given hoe such an icy tundra Antartica is, how exactly are researchers able to extract soil samples from ice? Also, what gave them the idea that the natural bacteria microbes from Antartica could be used to decontaminate oil spill in cold water climates?
The phylogenetic tree in this study was constructed utilizing a consensus sequence of the strains. The tree helps to identify certain pathogens that may be of close relation with one another due to ancestral links in DNA. What’s most interesting about this section is the degraders were hydrocarbon instead of PAH as in all of the previous paragraphs.
This is a great question. I too was wondering the possibility of BC1 being utilized in relation to Brucella would have any important correlating consequences for the use in health and agricultural uses.
This paragraph sums up the technique and nutrients utilized to prepare and grow cultures on NA plates. The plates contained diesel, pyrene, anthracene, phenanthrene, or a combination of the 3 PAH as the only carbon substrate. Results showed that certain nutrients and media provided different forms of growth. Isolates LC grew poorly on pyrene and phenanthrene, while LB grew quite well on all media except for pyrene. This test provided information that displayed the ability of the strains to grow a PAH substrates could possibly be useful for hydrocarbon bioremediation.
Find it interesting that rod-shaped bacteria are the dominant morphology, I would expect cocci or spirillum to predominate due to functionality and cross area. This article provides quite meaningful insight into proper function and shape.
The main idea of this paragraph is to inform the readers that certain bacterial inocula have quite specific recipients, and are unable to properly PAH-Biodegrade. It has been studied that bacterial inocula from the same region or similar areas have a higher percentage of effectiveness. Overall this particular study helps to identify, both genetically as well morphologically, the PAH-degrading bacteria from locations such as Eastern Province and Saudi Arabia to determine the limits of their degradation abilities on specific PAHs.
I am still struggling to understand the concept behind this procedure. There seems to be a certain factor behind PAH degraders that does not compute to me. How are the pellets utilized in this experiment?
I agree with Abrianna it is interesting how the difference in temperature can cause a significant difference. Also, what are the components within the cleanup kit?
I believe I see a recurring trend from another article where certain bacteria from the same location display have similar morphologies. In this particular case, they are all gram-negative and rod-shaped. My main concern/ question would be what causes the differentiation in the efficiency of the degraders being that they are retrieved from the same location?
I believe this is an interesting question, the fact that there is a lack of research in this field leaves room for more research to determine the full effects and possibilities for the use of deep water hydrocarbon degradation. Is this possibly a revolutionary discovery or one that can be utilized for decades to come?
How long has the SIP technique been utilized and are there any other techniques available to replace this one that can be more accurate? Also, with depth, do the levels of PAH’s intesify or become less severe.
I was wondering about this as well, I also didn’t see any information related to this topic. As for the microbial mats, I believe this would allow for a quicker cooling time.
I’m not entirely certain of this answer, but I believe it’s to keep the sample from becoming inactive or degraded by the light, which could cause forms of bacterial growth.
Of the 6 PAH’s tested, which would be the most likely to be utilized in future research as a plausible resource/ remedy to natural bioremediation.
I have a similar question, what were the criteria for the selection of the PAH’s and why were there not more than six species chosen for this particular experiment?
What acid was utilized to cause the oxidation to occur in 3 days, also; does this time frame suggest that this occurs at a slow, medium, or relatively fast pace?
Is there any particular reason why one band containing 13C enriched DNA was more dominant than the others?
What chemical or process causes the reduction of selenium oxyanions to produce a red elemental selenium?
Since Enterobacter cloacae are able to accumulate as extracellular granules, what effect does this have in the environment in which it is found?
What was the purpose of adding antibiotics to the medium. What effects would this have on the growth rate?
Because S17-1 was plated on an LB agar containing tetracycline, which is utilized as a marker for selecting colony vectors. What effect would sodium selenate have?
This may be a simple question but, what does the term cosmid mean when referring to the cloning procedure? Also, how do cosmid clones differ from other types of clones?
The sizes are characteristics of the different types of bacteria. This would help to differentiate the two cells during microscopy.
What is the significance of E, cloacae not being able to use Se(VI) as a sole electron acceptor for anaerobic growth?
This paragraph describes how the wild type fnr mutation was able to restore removed phenotypes. An FNR-like protein was found in E. cloacae which correlated to anaerobic transcription and are an essential protein for the reduction of Se (VI).
If PAHs are formed by two or more aromatic structural configurations, is there a configuration of these structures that is not hazardous to humans. If a person were to come in contact with PAH is there a way to reverse the effects? How long would it take for the symptoms to be reversed?
How many aromatic ring-hydroxylating dioxygenases are there in total? When referring to the multicomponent enzymes, the alpha and beta regions, what are the conserved regions on the alpha particle and how are they used? It’s interesting to learn that aromatic structures play a substantial fundamental role.
It is amazing to me how aromatic hydrocarbons can affect the world in ways such as fossil fuels. How they can be released into the marine environment causing forest or grass fires from petroleum by-products. The fact that PAH’s are cytotoxic, genotoxic, and carcinogenic to marine life and can be transferred to other organisms is interesting to learn.
What causes marine bacteria to be a more significant PAH degrader than the terrestrial habitats? Will there be a larger study on marine bacteria PAH degradation mechanisms? What amount of research was required to determine these facts about obligate marine bacteria, and since there has been limited research, what steps are needed to advance the study and understanding?
During the incubations @ 21 degrees Celsius, why were the 14CO2 mineralization levels inexistent for FLU, BaA, PHE, ANT, & PYR.
How might the results be skewed if incubation times were shorter? Also how did they choose 12 says as the proper time to gain results?
OTUs are used to categorize bacteria based on sequence similarity. Each of these cluster is intended to represent a taxonomic unit of a bacteria species or genus depending on the sequence similarity threshold
Why did they only use the ultracentrifugation technique to isolate heavier minerals for only the 13C model instead of the others as well?
Essentially, they used the acid to kill the bacteria, so when they incubate it, they are looking for how phenanthrene reacts with other sediments in the sample not just the bacteria. Correct ?
Pyrosequencing is a method of DNA sequencing based on the “sequencing by synthesis” principle, in which the sequencing is performed by detecting the nucleotide incorporated by a DNA polymerase.
It’s really fascinating that PAH-degrading microbes can have enhanced PAH degradation potential by the addition of inorganic and organic supplements. However, will these enhanced PAH-degrading microbes be used in bioremediation? What affect could the inorganic or organic nutrient supplements have on the environment as a consequence?
Toxicity of PHAs are dependent upon a variety of factors such as structure or the way they are exposed in the environment. Which PHA is worse depends on the context of your question. For example, in marine systems LMW PAHs are acutely toxic to marine organisms while HMW PAHs are not.
http://ceqg-rcqe.ccme.ca/download/en/243
It is interesting that authors of this study chose to use spread plate as their pure culture technique. Is spread plate more efficient than streak or pour plate under these conditions?
Several times throughout the paper it has been mentioned that cultures are “twice washed and resuspended” in their receptive media. What exactly does this mean? What is the purpose?
What makes Nap and Phe so resistant to degradation by Rhodococcus sp. CMGCZ? Why is Fla so susceptible to degradation by Rhodococcus?
Does the YE-supplemented medium stabilize degradation rates that results in a continuous degradation rate with no delay? What caused the sudden rapid phase of degradation on the MSM plate?
Paragraph 12 states that Rhodococcus sp. CMGCZ exhibits potential to degrade PAH Nap, Phe, and Fla as sole carbon and energy sources. Because of this wouldn’t Rhodococcus fit better into set one rather than set two? Despite degrading Fla more efficiently, Rhodococcus still has the potential to degrade Nap and Phe.
It’s really interesting that the YE supplement resulted in the absorption of YE by Nap rather than enhanced degradation. Why did this occur? Is there another supplement known to increase degradation of Nap?
I had a hard time understanding what was meant when the authors described rhodococci as “mycolate-containing nocardioform actinomycetes”. After Googling a bit, I think this means that rhodococci contain salts/esters from mycolic acids ,which are long fatty acids within the cell walls, and that they are branching bacteria that reproduce by fragmentation of filaments.
When would Rodococcus sp. CMGCZ be able to be used for bioremediation operations? Are there certain requirements it has to meet first before being introduced to a polluted site?
With chronic exposure to PAHs the most common effects are symptoms of various types of cancers such as skin, lung and bladder. It’s also interesting to note that its hard to attribute a single symptom to a single PAH because often PAH exposure is by a PAH mixture rather than a single type of PAH.
https://www.atsdr.cdc.gov/csem/csem.asp?csem=13&po=11
It’s really interesting that PAHs are often transferred to humans by the consumption of seafood. Don’t let this alarm you, the environmental protection agency has stated that taking in small amount of various PAHs every day will not cause harmful side effects. The FDA also has a method for detecting 15 types of PAHs and their concentration within foods using QuEChERS-based extraction and high-performance liquid chromatography.
https://www.fda.gov/media/114401/download
For anyone else who didn’t know, sonication is when sound waves are used to disrupt particles in a solution. In this case it is being using to disrupt the cell membrane and release the cells contents. Are there alternative methods for disrupting a cell?
Restriction enzyme digestion is used in order to create DNA fragments that can be ligated together. This is how DNA is prepped in order to be cloned.
Did the authors choose to screen for activity rather than DNA sequence or protein homology because prior knowledge is needed for the latter?
I think you are correct! The aromatic oxygenase gene is what encodes the aromatic oxygenase enzyme which oxidizes subtrates, in this case indole, by transferring oxygen to the substrate.
What affects the activity level of PhnC? What caused the activity of PhnCs with fused, unfused, and monocyclic catechols to the so similar despite having different structures?
The authors make sure to explicitly state that the order of the genes are different from those previously reported for aromatic ring dioxygenases. Can someone help me understand the significance of this?
What I found is that a flanking region is a region of DNA that has not been transcribed into RNA but is adjacent to one end of a gene.
What causes PhnC to be involved in both upper and lower pathways for degradation of naphthalene, phenanthrene and biphenyl? How does this effect rates of degradation?
From my understanding, FISH technique can be used to identify bacteria in environmental samples. Is it possible that this was the method used or a different one?
This is a great assumption! The PCR we did in lab likely did not go as planned due to the primers not fitting well enough with the genes to produce a PCR product. Maybe it is also possible that the reagents used for PCR were faulty or inappropriate concentrations for the master mix was used. I’m sure this will be elaborated on in the discussion section!
What do the authors mean when they say “all amendments of electron donors and acceptors were made by syringe injection from sterile, anoxic stock solutions?
I think it will be very interesting to see how the organisms will respond to varying temperatures and how it could affect oxidation/reduction.
I did a little research on Euryarchaeota and found out that although this phylum is often found in extreme environments they have also been seen in moderate environments as well!
I think when they author’s state that oxidation and reduction being potentially capable of cooccurence when incubated in light under anoxic conditions means that it is likely that these reactions are coupled as seen in normal redox reactions.
I think we see so few organisms not only because the environment is extreme but because of the size of it. Assuming the ponds they are sampling from are small, there would be a smaller population to reduce competition for resources.
This is what I gathered from reading this as well!
I believe the purpose was to confirm their suspicions about what electron donors could drive reduction.
It would interesting to see a similar experiment done but using the green biofilms rather than only using red biolfilm samples.
Since we already know that about regularotry genes from other strains of this specie, is it possible that these genes could be similar but just in a different placement in the DNA.
These are the same genes that were done in paper 1 that we read, are they using the mutated strain of A$ to see the degradtion of acenaphthene? Also The regulatory geen clustered tha they isolated, i thougth they already foudn the properties that it was involved in the 1st steps of degrdation of the PAH acenaphthene
what is a Gm? also why did they filter the membrane onto the LB agar? what does this do?
WHy were so many plasmids added cotrasnformed? I know they were looking for the particular gene expression, but why woudl they not just put the plasmids in there own ecoli and grow up separatly?
I believe that this is what is happening. That these strains did not have the gene able to intitiate oxygenation of the acenaphthene.
Since they determined where the 4 mutant mini-Tn 5, they used the strain that AG3-69 to look for where they were inserted into the gDNA? or that they were part of the gene encoding for the ferredoxi reductase?
this redA2 homologue is likely involved in the oxygenation of 1,8-NDCA or its metabolites in combination with other oxygenase components, which have not been identified.
is this the gene that they were analyzing in the results section like paragraph 4 i think? that deals with the mutagensis process? Also this gene is important in the oxigenation of the PAH in question, so it should be inhibited if the mutant strain is to not oxygenate the PAH.
In strain A4, arhR is located upstream from arhA3 in the gene cluster; however, the intergenic region between arhR and arhA3 is about 1 kb in length and includes putative transposase genes or their remnants (ORF8 and ORF9). Therefore, autorepression as with typical LTTRs does not seem likely to occur with the expression of arhR.
the LTTRs are not important in the oxygenation of PAHs, or the genes that are expressed to degrade PAH inital step. They are only expressed when the PAH is not present or the genes are not able to be expressed?
CJ2 has had multiple pathways identified in PAH degradation? Has any of the other strain in this paragraph as well? I just am confused on what it was saying a little bit. the genomic island, phn, it tell us about the catabolic pathway that PAH are degraded? is this what other research has been off of too? or did they follow up off the previous papers that we read.
Since this is a new strain, what other bacteria would Strain P73T be closest to when trying to identify the genes for PAHs degradation? Would it coming from the indian ocean maybe have genes that are different from ones already identified?
Why would they choose to do a mutation, if they don’t have the full genome, nor do they have any information of where the oxygenase genes are located on the DNA?
nevermind, i see where they did it
Why did they choose the cre-lox recombinant method? do they have a little more control on what DNA would be knocked out?
It seems that new genes were discovered for the PAH degradation, so will they have to due a knockout mutagenesis or out in them in a expression vector to make sure they are RHD genes? Does the P73T genes unlock more doors to the wide range of RHD genes that are out in PAH degrading bacteria?
These are 9 genes that are know, but does that mean that if these genes are not present, does that mean that other genes could be there, but the primer did not bond to them?
What is in the kan cassette? what is inserted into the genome of the P73_0346
Why is important that the transporter genes were located in the vicinity of the aromatic catabolic genes? Do they transport these genes? or is this due to like energy source? Also the transmembrane protein genes that were flanking, are these important for PAH degradation?
Why are knowing about these sugars important for the bacteria? Is this for growing up on like a TCA test, which it can use better? I thought glucose would be the best. Also since it is known that they can use these types of pathways, how does this help in the PAH degradation?
[The P73T genome contains 138 candidate genes that may be involved in the metabolism of aromatic compounds, including genes that encode six ring hydroxylating dioxygenases, eight ring cleaving dioxygenases, other catabolic enzymes, transcriptional regulators, and transporters in the degradation pathways.]
Why was further testing not done to help understand these other genes, especially the ones in regionB? I feel like it would have helped with understanding of these newly discovered PAH genomes.
Why did they not isolate the 5 plasmids and do more work with them? I understand it is a genome paper and they discovered some novel genes dealing with PAH degradation, but wouldn’t these plasmids be novel in understanding why these genomes were so important in understanding PAH degradation pathways? I just done understand why they didn’t go any further in their research.
The dissolved selenate was analyzed by ion chromatography. is this method the most efficient for observing this or would an array of methods provide more accurate results?
Isolation by sodium dodecyl sulfate lysis is a form of isolation I am not familiar with. How does this method work and what is the advantage of this method?
The effects of PAHs in the human body is interesting due to their environmental threat. I wonder if the way that PAHs enter our body effect the long term disease that we could experience.
Bioremediation is one of the most interesting uses of microorganisms in my eyes. The positive use of microorganisms to degrade PAHs I’m sure has a huge effect on the diseases that can be caused by PAHs.
I wonder if letting the plates incubate for two weeks instead of 48 hours had any effect on the bacterial colonies exhibiting zone formation. It is interesting that they only incubated CMGCZ for 48 hours after it was picked from the MSM plate, but allowed it to incubate for two weeks to form the zone colonies.
Why was the run a degradation test only in YMSM in different concentrations. Would it make a difference if the degradation test was run in MSM?
I found it interesting that a delay was observed in Fla degradation for the MSM plates, but not the YMSM plates. I think more information on the chemical properties of the agar plates would help determine this trend.
The difference distinguished between Fla, Phe, and Nap was the opposite of my original thoughts. I thought I would see the exact opposite trend. It is also interesting that the MSM and YMSM plates experience the same difference in degradation.
After reading this entire study and the looking at the molecular weight and chemical composition of Phe, Fla, and Nap, it is still unique to see the degradation potential of Fla compared to the others. Not only would I have hypothesized the results to be flipped before reading this, but I would at least expect the results to be less drastic than what they are.
Looking at the effect yeast has on the degradation potential is interesting to me. Even though the degradation potential of ILCO was not that high, adding yeast as a carbon source still aided in the degrading potential. The results of adding yeast were also experienced in the Fla sample.
With the diversity and capability of rhodococcus, I would be interested to see how its enzymatic effects could benefit humanity. If applied in a pharmaceutical setting, it could make multiple breakthroughs.
I wonder why the substrate was found to not be essential in the degradation rate in the Rhodococcus strain but essential in the R. erythropolis strain. If different substrates were used what would be their effects of the diesel fuel degradation?
If PAHs have the potential to cause cancer, I wonder how often marine life has been at the root of what initiated cancer growth in a human. Are there tests performed that takes away their toxicity before marine life is served to customers?
I’m interested to see the effect of studying a marine bacterium instead of a terrestrial bacterium. I wonder if using these different PAH degraders will have an effect on the overall efficiency.
Determining the function and structure of this set of genes in the Cycloclasticus sp. strain could be critical to determine what genes have significance in the degradation of PAHs. This could allow for the determination of other effective bacterial microorganisms that could be used in the degradation of PAHs.
Interpreting this data and reviewing the methods have helped me to understand the meaning of our work in lab. I now feel more equipped and driven to evaluate my data knowing that our lab work has significance in other experiments.
I wonder if they thought about seeing the effects the chemical purity had on the experiment. I would be interested to see how the purity would effect the overall outcome. I’m assuming it is a significant detail considering the author made a point to mention it.
After doing some research, it was interesting to find out how vital the choice of media is the experiments. We know that ONR7a media is critical for naphthalene degraders because it uses naphthalene as its sole carbon source.
I wonder if the fact that both constructs had a 10.5-kb Sau3AI fragment contributed to the results of both constructs containing an aromatic oxygenate gene. It’s also interesting to me that what we are learning in lab is applicable to these experiments.
I find it interesting that sequencing genomes allows us to see the metabolic properties of these ORFs. I wish the author would’ve told us whether or not this ability to sequence correlates to a specific sequence that can degrade PAHs.
Looking back at the introduction of this paper gave me the thought of if terrestrial bacteria versus marine bacteria had any significance on where the gene clusters were localized. Does the fact that the Pseudomonas sp. strain is a terrestrial bacteria have any influence on its plasmid localization?
What is the difference in the alpha subunit and the beta subunit? What percent similarity was the beta subunit if they both fell outside the major cluster?
I’m interested to see how the outcomes of this study are given. Considering this study will help to further understand the evolution of arsenic from anoxic to oxic conditions.
I’m interested to see why they were not able to obtain PCR products and if it had any correlation to the primer or the enzymes used in the reaction. I’m hoping the materials and methods will reveal what enzymes were use to produce a PCR product.
It is so interesting to me that after reading this entire paper to think about how critical metabolic reactions such as the ones with various arsenic species are in the development of the oxic environment we have today.
Why was it noted that the slurries were stored in the dark? Was the conditions of it being in the dark and held at 5 degrees celsius the reason it was kept viable?
I wonder why they used a permeable foam stopper. I wonder how it would effect the growth if there was no exchange with the atmosphere.
Were the photosynthetic bacteria not able to survive in the green-colored springs? Is the temperature to high for them to survive?
I think it would be interesting to look at the Eurarcheota phylum of Archaea and look at the similarity between the Archaea and the Bacteria found. Maybe they have similar components that could benefit other studies.
It is interesting to see how the oxidation and reduction rates lower each times the light was reimposed on the sample. I wonder if the amount of light used to control this experiment varied at all each time it was reimposed.
It is fascinating to me that in all of the papers we have looked at this semester, the 16S rRNA is foundational part in building a phylogenetic tree. The discovery of the endosymbiotic theory and the conservation of the 16S rRNA has impacted biology tremendously in making these biological connections.
If acetate was used as an electron donor in this study, why was there not an equal and opposite reduction.
What are some of the most effective types of bacteria that are used in the bio remediation process of naphthalene?
what are some of the physicochemical treatments used if biological methods fail?
I would like to see if other environments such as the Caspian sea or the Mediterranean would yield different results.
is the glycerol just enhancing the growth of the different bacteria? or what purpose does it serve?
what is the flame ionization detector? what is its role in isolating the naphthalene degrading bacteria?
Was there an underlying reason for 15 days and 200 ppm of naphthalene chosen as the growth conditions in the experiment?
As a whole what caused all the naphthalene strains to ultimately decrease overall concentrations when they were grown at greater than 400 ppm?
Could the introduction of these multiple bacteria strains have any effect on nutrient levels? or marine life? Like eutrophication for example? or would there not be enough of them to cause an issue?
It is mentioned that the Persian Gulf has a high diversity of naphthalene degrading bacteria. It seems like there is a evolutionary reason for the high diversity due to all the oil traffic interactions in that area.
It might be an environment with not as much pollution, or possibly just lighter pollutants that don’t contain as many “heavy” compounds.
what results from the previous study were so compelling to override the extensively studied terrestrial habitats?
specifically for the strains that grow on naphthalene. would some of the bacteria we isolated in the previous paper be useful? Since they are naphthalene degrading bacteria and this genus of cyclolasticus can grow on naphthalene.
apparently the Luria-Bertani agar is one of the most widely used media for bacteria. What makes it so commonly used?
what does the ferredoxin fragment help us with in this experiment?
Why was the ferredoxin fragment amplified? I know its an electron acceptor in photosynthesis but what role does it serve in this experiment?
Does the size of the clusters of ORF’s have any correlation with its degradation ability of PAH’s? I see size related to each other but not efficiency compared with one another.
why does the change in position of the 3-methylcatechol as opposed to the 4-methylcatecol cause such a change in activity? Obviously the addition of a methyl group increases the relative activity for the catechol substrates.
what does a partial ORF consist of? I guess visually I don’t see how this isn’t the 10th and 11th ORF? Does it have something to do with the start codon specifically?
it is interesting that the phnA was able to transform fused cyclic compounds but also unfused ring structures as well. Biphenyl and diphenylmethane are examples of the unfused compounds
what do they mean by “upper” and “lower” pathways of degradation?
apparently the diel cycle or “vertical migration” is a pattern of movement for marine organisms.
It goes much more in depth than simply that but in essence during the day these microbial mats respond differently than they do at night. This has to do with the presence of more oxygen providing more oxic conditions. Where as at night photosynthesis ceases and the mat becomes more anoxic and levels of H2S increase. The organisms respond to the change of conditions and rise or fall.
It may have something to do with the photosynthetic activity of the biofilms.
so I didn’t know what the nylon filters were used for so I looked it up and apparently the nylon filters provide extra stability for the HPLC sample preparation.
what causes the oxic test to yield the same result as the anoxic test?
I’m interested in what the percentage of acetate was for the 5mM As(v) + 3mM sulfide. Table two says this number was not determined. ( I guess that is what ND is standing for here). I’m Interested because these additions had the highest bicarbonate assimilation numbers. What made the acetate assimilation percentage not able to be determined?
no it would not seem to be involved. That makes more sense now thank you.
what about the dark As(V) reaction made the temperature range broader than the light induced As(III) reaction?
So the halophilic nature of the related Archea would not be present if the clones were take from an area with less salt concentrations? Is that what they are saying?
what does it mean by “primers were not quite suited for this particular environment”. How can high salt concentrations cause this to occur?
It interesting to me that something as simple as yeast can help degrade the toxic PAHs. The fact that we find such important usages for such small and seemingly insignificant organisms, like yeast, to help clean our ecosystems is remarkable to me.
What are the differences in the inocula from the different locations? What makes the ones from nearby locations better than ones from far away locations?
Do I need to be able to understand the steps they have described? This section went way over my head. What am I supposed to take away from this section? Because it is bogged down with details I don’t understand.
Doesn’t temperature affect the structure of many key players in biological processes? The main thing that comes to mind is the denaturation of proteins and enzymes at different temperatures. I don’t know if that is what is in play here but it could be along the same lines?
Are they only able to tell the rod shape by looking at the SEM images or is it visible on the agar plates as well? I looked at figure two and the only way I can tell that they are rod shaped is by the SEM images.
What would show that P. aeruginosa is an opportunistic human pathogen? What would be seen on the genomic studies to indicate this?
So the production of a bio-emulsifier makes the microb a better PAH degrader than just producing a bio-surfactant? Is that for all PAHs or just the high molecular weight PAHs?
In a solution where evaluating the turbidity would show the change in colony population, the solution would be cloudier the colony population increases. Is this only for colonies that are not PAH degraders?
I was confused in the difference between PAHs and naphthalene. I was confused because they seemed to be the same thing but were also used in different parts of the introduction. I googled it and it was cleared up that naphthalene is a type of PAH. It is what is used to make mothballs. It is a good PAH to use in research.
Is the upward migration of the petrochemicals similar to the marine oil snow that occurred after the Deepwater Horizon explosion? Is that what makes this a good model system to study?
what causes the temperature differences of the samples? is it due to the differing sulfide concentrations?
Does the warmer temperature of the second sample allow the microbial mats to form or is it the microbial activity and presence that makes the second sample warmer and have hydrothermal influence?
So the 13C is what they want to focus on and they tracked the 12C and 14C as reference points to help understand the 13C values?
So with all of these genera being PAH degraders, they are found naturally in the deep-sea environment. Are these studies leading to how to increase their PAH degrading capabilities/how to make them more effective? With them occurring naturally and you stating that this means that we don’t have to create superbugs, are we just trying to increase their capabilities?
Is it better for the bacteria to be obligate or non obligate? I am thinking obligate because that means they always degrade the PAHs but I don’t know if I am correct in thinking this way.
So just the location of a gene can change the way an organism functions even though it is the same gene as another organism? I didn’t know the location of a gene could be responsible for so many differences.
What is a prosthetic constituent?
So the gene that controls the reduction of selenium is the regulatory gene fnr, but the different forms of reduced selenium are due to the differing enzymes used in the reduction? Does this mean that every selenium reducing microbial has the fnr gene and then different enzymes contribute to the reduction in different organisms?
So three strains were tested and two of them were found to reduce selenium but the genomic library was created for the strain that does not reduce the selenium?
Because we are looking at transconjugants, am I right in saying that this whole experiment is about the selenium reducing gene and not the ability of the bacterial strains to reduce selenium? I want to check because the previous papers all looked at the different bacterial strains doing the degrading.
So to show that they are degrading selenium, they just look for any type of selenium particle that would have been formed from the reduction of selenium? Is there a specific particle they looked for or just any selenium byproduct?
In the LB agar, would the sodium selenate make the agar a differential medium giving the colonies the red or white color depending on if the colony could breakdown the sodium selenate?
so the bacteria will not reduce the selenium until all of the oxygen is used up. is this because it will use oxygen as the electron acceptor until it runs out and then it will start to reduce the selenium by switching to it as the electron acceptor?
So would E. coli be able to reduce selenium if it had the oxygen-sensing transcription factors? is that the only thing holding it back from reducing selenium?
I do agree with your statement about oil being a natural resource that will not be given up any time soon. The paper says, “oil spillage due to transport, storage, and utilization of fossil fuels has become a serious and persistent threat”. So, I think the way we can limit the amount of contamination thereby having no effect on the ecosystem, is if we can find a different way of transporting, storing, and accessing the natural resources that result in less or no spillage. I’m not sure if this is possible since we haven’t figured it out yet. Of course, whatever methods we can resolve must take into account the cold weather and extreme winds of the Antarctica climate.
Hey Abraham, from what I understand after reading an article on Towing Icebergs from Antarctica to the United Arab Emirates in 2018, is that they aren’t sure “exactly how water would be extracted from the iceberg once it reaches the UAE, though the general idea is that the icebergy will be cracked into many pieces, chiseled off bit by bit and loaded into tanker ships which would take the ice to shore and the liquid water that results from the ice melting could then be purified and sold to water companies and government agencies.” Furthermore, 30% of the iceberg would melt during the towing but the amount available once arrived would be between 100 million to 200 million cubic meters of water which is enough to provide around 1 million people with drinkable water for a total of 5 years.
Hey Ryne! From what I understood from the introduction, diesel oil contains PAHs like phenanthrene and it is through bioaugmentation that the researchers “directly seed” those contaminated sites with pollutant-degrading bacteria, i.e. PAH-degraders resulting in what they’re trying to accomplish: bioremediation, which they described as an “economical, and safe approach that can be applied to the decontamination of PAHs with minor alteration of the soil”.
Hey Kamryn! I was also wondering the same thing. I understood that E. coli was used as a negative control as E. coli is unable to grow in either condition. However, it struck me with surprise that there was such a difference in the 2 graphs: with R. erythropolis and S. xenophagum growing more CFU/mL with 0.2% diesel-fuel source vs 0.05% phenanthrene as sole carbon source. Furthermore, P. guineae was extremely drastic in comparison with success in growth using phenanthrene as the sole carbon source but not using diesel fuel.
What would be the significance that the “possibility exists that other strains of interest in our soil samples were not identified”? Are they just saying that since their main goal was to isolate the PAH-degraders, they felt the need to clarify that it wasn’t completely isolated?
[Novel techniques of in situ microculturing have been used to culture Antarctic soil bacteria, by means of a soil substrate membrane system (SSMS), which recreates the native conditions of growth by using the collected soil samples as the nutrient source. ]
Is this similar to a continuous culture or would it be a batch culture or something different altogether? I could be making connections where there are none. I would think it is a type of continuous culture because they didn’t say anything about an accumulation of wastes like in batch culture. Continuous cultures are also better than batch cultures because you can manipulate the amount of nutrients and dissolution rates which would seem beneficial to do.
[The remarkable finding that atmospherically-transported Saharan dust enables proliferation of vibrio bacteria by delivering dissolved iron to surface marine environments]…
This is really cool because in my sedimentology class, which is a branch of geology that deals with the life cycle of sediments and sedimentary rocks, sediments from across the world can travel a really far distance and end up in the oceans through a process called erosion. I never thought about the different heavy metals, like iron, that could be transported along with those sediments.
I noticed in this paragraph (#7) that they state that gram-negative bacteria use a membrane-spanning transporter protein, a siderophore-iron complex, as well as an inner membrane ABC transporter all for the purpose of iron uptake. This was very interesting to me because when we were learning about nutrient uptake and transport in CH. 4, I was under the impression that those processes (ABC-transporters, siderophores, coupled-transport systems, etc.) were separate/independent from each other. I did not know these processes could employ a series of events and co-regulate together.
[E. coli S17–1 λpir was used for cloning, and E. coli MG1655 was used for heterologous gene expression]
I was curious as to what E. coli had to do with experiments involving V. harveyi and V. fisheri, specifically why they used E. coli for cloning and heterologous gene expression, but then I remembered that E. coli is a common model organism in the field of microbiology. I googled the specific reasons as to why E. coli is a model organism and the website cited stated that E. coli has a very simple genome, a fast growth rate, and among other reasons, it is a well-studied organism. Even different strains of E. coli provide substantial contributions which makes sense as to why they are using one strain for cloning and another strain for the heterologous gene expression.
What I understood from this paragraph was that they are using reporter genes to identify whether or not the gene iucABCD is expressed, as this is the function of reporter genes. The fact that it is fluorescent makes it easily detectible in culture. However, will the fluorescence also be easily detectible in the PCR and gel electrophoresis results?
What does it mean that culture fluids containing V. fischeri prevent the growth of vibrio species in minimal marine medium? From what I understand is that V. fischeri is able to uptake the limited nutrients quicker than other vibrio species, consequentially leaving no nutrients left. I also understand that the vibrio growth inhibition only occurs when V. fischeri is added to the culture fluids.
[V. fischeri ES114 culture fluids only inhibited V. harveyi growth when V. fischeri ES114 was cultured in minimal marine medium but not when it was grown in rich medium]
Why is the growth inhibition only present in a minimal marine medium and absent in a rich medium? Is this because the V. fischeri inhibitor molecule works better in a limited nutrient medium whereas, in a rich medium, it is easier for vibrio species to acquire nutrients with or without the inhibitor molecule?
Hmmm, this makes sense because the evolution of siderophore production, i.e. specialized molecules secreted to bind ferric ion and ultimately being reduced into a more useful ferrous form, have contributed to V. fischeri’s acquired competitiveness in its surrounding environment by leaving no iron to the other species.
In paragraph 5 which is what I am thinking Dr. Níhain is referring to when she says “read on. they go through some possible explanations…” they explain that having the AerE gene allows (1.) immunity to cytoplasmic aerobactin toxicity for the cheaters; and (2.) enable aerobactin recycling by the cheaters allowing iron accumulation and acquisition thus increasing their growth.
It seems that the main issue this paper will be focusing on is the genes and enzymes related to aerobic ammonia oxidation, the “first and rate-limiting step of nitrification”. The introduction states that ammonia monooxygenase is one enzyme found in aerobic ammonia-oxidizing bacteria, which is important to the overall research because it is also available in an MGI crenarchaeon, which are likely important and substantial nitrifiers in oceans.
Because of the lack of studies on nitrification along oxygen gradients, this research is being conducted within the Black Sea as the Black Sea is the perfect model. They are trying to identify the marine species responsible for the upkeep of nitrification processes in areas where remineralization, leading to ammonia production and anammox, does not occur.
I think they are analyzing the anammox rates by pairing isotopes because of the fact that isotopes can be used as “bioindicators” and help map out different sources and states of nitrogen (in this case). Since the purpose of this article is to understand better the coupling of nitrification and denitrification/anammox, it makes sense that different isotopes of N are analyzed.
From the PCR they might expect to find evidence of the 16S rRNA in order to determine if the amoA gene is present. This will help them narrow down the nitrifiers within the collection sites of the Black Sea and help them understand the coupling between nitrification and denitrification.
Nitrification levels are basically nonexistent in the anoxic zones and tend to increase in the suboxic zones and then decrease in the same fashion in the lower oxic zones. Furthermore, anammox bacterial cells show a wide variety of activity in the suboxic zones and very little activity in the lower oxic and anoxic zones. As stated in the introduction, substrates from nitrification (a) are used for denitrification processes (d), so it would make sense that A and D have this relation (i.e. as nitrification levels are low, anammox levels are high and vice versa).
(A) shows a high prevalence of crenarchaea cells containing the amoA gene, but a low presence of mRNA copies in the suboxic zone.
(B) shows a low prevalence of the BAOB cells and the mRNA copies in all 3 zones
(C) shows an increasing trend of YAOB cells in the suboxic zones and an alternating increase/decreases trend of mRNA copies between the suboxic and lower oxic zone.
This makes sense because as stated in the introduction, nitrification and anammox have been reported in the lower oxic and suboxic zone, respectively.
[Most of the obtained sequences fell into the marine clusters A, B, and C (20), but three OTUs fell into the “sediment” cluster, which also included Candidatus “N. maritimus.”]
What is the significance of classifying marine clusters vs sediment clusters? I would think the sediment clusters would correspond to deeper depths due to sediment properties–thus, located in 110 m or deeper (i.e. the anoxic zone which is not correlated with nitrification or anammox).
With the help from the legend, when comparing amoA genes found at depths of 80m vs 100m, you can see that there is a majority of species within the phylogenetic tree with amoA genes corresponding to depths of 80m. Furthermore, there is only 1 species with the amoA gene corresponding to a depth of 110. It is interesting that the only species with the amoA gene that has been observed at a depth of 110m has also been observed at 80m and 100m: that species being BS160B4.
I think the purpose of isolating the PAH-degrading bacteria goes back to the introduction where it is talking about microculturing. “New techniques of in situ microculturing have allowed to culture an even broader number of PAH-degrading organisms by recreating native conditions of growth, allowing growth of bacteria not normally isolated using standard culture conditions.” This is essential because they are trying to find the most effective bacteria strains for bioremediation, i.e. the process of safely, economically, and ecologically degrading those harmful PAH compounds.
The purpose of using fluorescence spectroscopy is due to the unique ability of PAH compounds to be fluorescent as stated in the introduction. The use of this type of method of visualizing microbes is better than light microscopy because LM emits a higher resolution allowing you to see the molecular dynamics and interactions in 3D. Thus, if you only used a light microscope, you would not be able to correctly or efficiently quantify the PAH compounds.
Woops, my last comment on fluorescence spectroscopy was supposed to be in regards to paragraph 4 on quantifying phenanthrene.
Could microorganisms that intake energy from hydrothermal vents help with the biodegradation of oil? If so, this would be an efficient and clean way to help with this problem
Salinity/oxygen levels are differ greatly from the surface of the ocean to the floor. Data samples would be more accurate if they were collected from the floor and surface so that way you could test the differences.
In what way, specifically, does Naphthalene hinder mitochondrial respiration? Are the effects on the mitochondria permanent or once the Naphthalene is no longer in contact with the organism, does mitochondrial respiration resume as normal?
What was the result of the bioremediation in the Persian Gulf? Were the majority of the PAHs bioconverted into micorbial biomass, carbon dioxide, or water or just a fraction? Is there even a need to bioconvert all the PAHs? or will partial conversion be sufficient to make the Persian Gulf “safe”?
Were only the seawater samples transported on ice? or were the sediment samples also transported on ice? Is there a significant reason why the sediment samples were taken at a shallower depth range than the seawater?
Is there a reason why smaller amounts of seawater portions were used as opposed to portion sediments?
What happens to the bacteria when their food source is no longer available? Without a food source would the bacteria just die or would it be possible for them to mutate to obtain food from other sources?
Have there been studies done that prove the resulting biomass is completely benign? What effect does the resulting biomass have on the ecosystem of the effected area?
What was the purpose of filtering the seawater before adding it to the medium? I could see maybe filtering out solid sediments, but if they add the paper back to the medium, which would also transfer the sediments.
If scientists are using bacteria that are already present in the environment how will changing the concentration of those bacteria effect the ecosystem that is present in these regions?
So, it wouldn’t matter if the bacteria had a high
Emulsification activity if it had a low BATH%, since the bacteria wouldn’t be able to adhere for long enough to degrade the naphthalene? Am I reading this correctly?
Would more research need to be done to find strains that can degrade at higher levels? I don’t know the average ppm for oil spills, but I would think that, at least at first, the levels might be higher.
What is the concentration of naphthalene in the Persian Gulf? If it is higher than 400ppm what would the effects of releasing this bacteria on a large scale be?
Would each place have it’s own strain of these bacteria, or would there be similar strains in different places?
How does the process of bioremediation, specifically the introduction of large quantities of bacteria, effect the environment? What are the possible side effects of bioremediation in terms of possible decline of other types of bacteria that may be useful to other organisms?
Are all of the PAH degrading bacteria of the same species, or are they different depending on where the pollution occurs? Are all the PAH degrading bacteria close to each other on the photogenic, or have they evolved to be completely different depending on the environment and pollution encountered?
I don’t think they were measuring degradation of phenanthrene, just the bacteria’s ability to use it as the carbon source. I think they were double checking that they hadn’t isolated autotropic bacteria by mistake.
Were these tests done to ensure the bacteria were only able to degrade the phenanthrene? Wouldn’t the bacteria be more useful if they could degrade multiple PAH compounds?
I did a little research, and found out that S. paucimobilis is a pathogenic in humans. How would this affect the viability of using this particular bacteria for bioremediation?
I find it interesting that the bacteria fixed the phenanthrene more quickly at higher concentrations than at lower concentrations. I would think it would be the opposite because lower concentrations would mean there is less to fix which should make the process faster.
What would be the significance of microorganisms being able to use arsenic for energy? Are there areas where arsenic pollution is a problem? What effects do these microorganisms have on humans?
Aren’t the red-pigmented biofilms usually harmful to other marine life and the ecosystem at large?
Why is this particular PAH the model for high weight PAH degradation? Is it strictly because there are so many bacterial that are able to degrade it?
If there are no PAHs available, are these bacteria able to utilize other sources of carbon? I wonder if these bacteria evolved the ability to degrade PAHs as a response to pollution, and if after the PAHs are degraded what the possible consequences of large amounts of potentially pathogenic bacteria in the environment would be.
I would think that they used artificial sea water to ensure that the bacterial strain would grow without possible contamination by other species of bacteria that may be free floating in natural sea water.
I would think selective but enriched. Since it has the sea water in it, and not all bacteria are able to live in environments with a high salt content. It’s definitely a complex medium since we don’t know for sure what exactly is in “yeast extract”
I find it interesting that the strain that they were looking at has the genes for motility but those genes are not expressed in this particular strain. I would think that would be because in water they could just float to where they want to go. It may not be as effective as using a flagella, but it is less energy using for the cell.
I wonder if the strain of bacteria mentioned here doesn’t have PAH degradation genes because the pollution in the Arctic Ocean is not as widespread as in other oceans.
I know there is still pollution in the Arctic Ocean, but I would think it would be minimal compared to, say the Gulf of Mexico. Strains in the Arctic wouldn’t be as exposed to PAHs so the expression of genes for PAH degradation would not be there.
So the bacteria wasn’t able to use Tyr or was the bacteria unable to produce Tyr, meaning it would have to come from the environment?
So would that mean that the bacteria was able to cleave the phosphate groups in a different way?
It would be interesting to see what would happen if only the plasmids were knocked out, or if the plasmids were transferred to a different strain of bacteria. The plasmids may not encode metabolic genes but there has to be some sort of use or help that the plasmids give.
I would think having more pathways to use PAHs would be beneficial to the bacteria, since that would allow for degradation of more PAHs. This doesn’t seem to be a problem for this strain though because it says that this strain can degrade many different PAHs.
I agree with Lisa, it’s interesting that they are researching foreign bacteria. I would think that the foreign bacteria would have a larger effect on the ecosystem than using bacteria already present in the soil.
Abiotic stress is the negative impact that non living things have on the environment. This is saying that there are some bacteria that are helpful to plants because the plants are symbiotic hosts to the bacteria. The bacteria get a place to live and things to eat, and are beneficial to the plants by increasing tolerance to pollution in the environment.
I think that the researchers are saying that in this study the concentration of copper did not significantly impact PAH degradation.
I think that this paragraph is saying that the isolated bacterium is able to resist the normal damage that copper causes.
I would think it would be advantageous for the organism to have the PAH degradation genes spread through out the genome, rather than all controlled by one operon. That way, the organism would be able to produce different enzymes to catabolize PAHs, rather than just one.
When looking for the genes for PAH degradation, is comparative genomics the most useful part? Otherwise how would they even know where in the genome to start looking?
What happens when the PAH isn’t all the way degraded by the bacteria? What happens to the intermediates when the cell can’t act on them? Are they just expressed as waste by the cell?
Why were the looking at the water soluble-products? I thought that when PAHs were broken down by bacteria for carbon and energy they were completely degraded?
Why were they looking at overexpression of this protein? I think I’m missing something here.
Why was this particular strain picked to do experiments on? It wasn’t one that the researchers had isolated in nature. Was it previously proven to be able to degrade fluorene?
Does this mean they kept repeating the experiments until they figured out what the nucleotide sequence of the gene they were looking for was?
They cloned the genes into the plasmid to be sure those were the PAH degrading genes they were looking for, correct?
By doing this test were they looking for the protein used to degrade PAHs?
So, in this paragraph they’re saying that the primers originally picked didn’t work in this particular strain. But they picked the primers based on research done on strains that are similar.
So the researchers found a new/novel enzyme in this strain that catabolizes the start of the degradation pathway?
Is there a possibility that there are less harmful pathways for carbofuran degradation? I understand that’s not what they’re looking at here, but was just curious.
So this paper is reporting on the genes for carbofuran degradation as well as purposing the pathway?
In this paragraph they are discussing how they came up with the proposed mechanism for carbofuran degradation, right? Were they testing for intermediates in the pathway?
So they used a plasposon approach (among others) in order to find the genes they were looking for?
So they’re saying that the carbofuran degrading genes are expressed all the time, not just when carbofuran is present.
So in these mutants with the insertion, the bacteria were not able to use carbofuran as the carbon source, at all?
What would be the purpose of the pathway being constitutive? Is carbofuran that prevalent in the environment?
This would seem to suggest that the genes were acquired by horizontal gene transfer, right?
So, if the cfd and cft genes aren’t on the plasmid, then they would be on the chromosome, correct? That would suggest that these genes play an important role other than just metabolizing carbofuran, otherwise it would be energetically expensive to express the genes all the time.
How would it be possible for the bacteria to mineralize carbofuran but not grow on it?
How would researchers involved know that the bioremediation process is an “environmentally benign clean-up?” Are there tests that have proven that the bioremediation process doesn’t potentially cause more harm to local marine life of the Persian Gulf? I googled possible negative effects of bioremediation and had a hard time finding an answer to these questions.
How would you determine that a certain bacteria was capable of degrading PAHs for a certain environment?
Why did the sediment samples and seawater samples need to be transported on ice?
Why is the temperature profile for PCR brought down to 54 degrees Celsius and then back up to 72 degrees Celsius?
Why would the N1 strain have a lower BATH % than the N7 strain?
Why would N7 show maximum growth at 400 ppm if the other strains were showing dramatic decreases in growth at this concentration?
Should other zones have been considered for sampling that were not as heavily polluted?
Why would this have been the first report of these genera from the Persian Gulf? Why would this isolation not have been done before?
How could this research experiment be further carried out? Are there other naphthalene degraders they could consider?
What type of isolation method would you use to ensure the uptake of Gram positive bacteria?
Is there a way to test marine animals for PAHS before they are consumed by humans?
I find it interesting that PAH degradation mechanisms of obligate marine bacteria have only been partially studied. Is there a reason for this?
What is a Gigapack IIIGold packaging extract?
What is a Gigapack III Gold packaging extract?
What does it mean if something is subcloned?
Why would the oder of the phn genes be different now?
What could be the reason that no small subunit gene was found in the flanking region of phnAlb?
What could be a potential catabolic and evolutionary benefit resulting from multipurpose electron transport proteins?
Why would they decide to just focus only on the cycling of arsenic under anoxic conditions?
What is a diel cycle?
Why would they not have also determined sulfide by high-performance liquid chromatography?
What is the significance of incubation at 43 degrees Celsius?
The text states that there were large errors associated with the sulfide-amended samples due to variable kinetics of formation and destruction of various thioarsenic intermediates. Why would these factors have affected the reduction potential of sulfide more than the reduction potential of hydrogen?
I’m confused on why the identity numbers (86/87) for the dominant group (SDT-arrA-13) are significantly smaller than the other Representative clones listed.
I don’t understand why the acetate amendment would not have stimulated the reduction of As (V).
Is this paragraph saying that the arsenite oxidase genes related to the aoxB amplicon could still be there even though they were not detected?
Traditionally, lay people view Antarctica as a continent devoid of life. Why would we expect to see bacteria capable of metabolizing diesel fuel and other pollutants in such a desolate landscape? It is easier to understand why bacteria in a place like Nigeria would be more capable of dealing with pollutants, but why would bacteria in Antarctica develop these unique metabolic pathways? I don’t think it would increase their fitness in any way.
I would think that if we are considering microbial bioremediation, the soil would already be beyond natural repair. Would you not expect to have an irreparable change of soil ecology if you are adding a non-native species to tainted soil in the hopes of making it more habitable for native species? I would expect microbial bioremediation to take an extended period of time, and during this time, do we expect horizontal gene transfer to permanently change the gene pool of the native bacteria species?
I understand that bioaugmentation could achieve the same results as introducing a non-native bacterial species to Antarctica in order to deal with pollution. But wouldn’t bioaugmentation bring up the same concerns as introducing a non-native species because bioaugmentation is an inherently unnatural process?
After reading Zachary’s comment, I was curious about the choice of medium. After a colony was successfully grown on M9 minimal media, wouldn’t it be desirable to grow additional colonies from the colony that survived on the M9? Would a more favorable media be chosen for quicker proliferation? Following this line of questioning, is this the next step of the experiment? As I understand it, the goal of this experiment is to isolate diesel fuel degraders to clean up human caused pollution on Antarctica. If a more favorable media was then selected wouldn’t you get more diesel fuel degraders?
Based upon my limited experience in quantitative analysis, multiple extractions are more efficient at removing more of the desired analyte. To answer your second question, I do not believe that the more concentrated analyte should have any measurable effect on increasing the excitation-emission spectra of each sample. I believe the purpose of the excitation emission spectra is to confirm the identity of the excited analyte not how much is there.
The overlap between biochemistry and ecological niches is a very interesting topic to study. Usually when I think of an ecological niche I tend to see it as a place that a species (typically a multicellular animal) occupies and tries to defend from similar competition. It is strange to think of the unseen wars that bacteria wage against each other using the tools of chemistry to outcompete their competition. In many ways it is a much more straightforward mode of competition because of the tools that they use to survive.
I’m curious about the rate of return of the siderophores to their microbial producer. If ferric iron is only scantily present in the culture, as it is in many areas of the ocean, it might be a disadvantage to overproduce siderophores. The wasted energy on the production of siderophores must be putting a severe stress on the vibrio.
Thank you for asking this question. It seems like a great way to get comment points for the most confusing section of the paper. qRT-PCR= quantitative real time PCR. My guess is that used this technique to calculate how much RNA was in the sample. In short they wanted to find out how long the gene was that coded for the selective advantage of V. fischeri.
I’m not 100% confident with this answer but I think that they wanted to stop the growth of the V. fischeri was so they could put it in culture with a polymyxin resistant strain of V. harveyi. In normal conditions V. fischeri outcompetes V. harveyi.
I think it is interesting that Photobacterium angustum was unable to grow in the V. fischeri growth media. It’s strange how a chemical attack by V. fischeri was able to affect multiple different species.
This was the experiment that I worked on in class. I was originally hesitant to post a comment on this section because I did not want to repeat what was stated in the description. My groups take was that figure A shows that the complete deletion of the aerobactin operon would show that no aerobactin would be produced. The more important of the two figures is figure B. Figure B is further proof that aerE is the gene responsible for transporting the siderophore outside of the cell. When aerE is deleting the siderophores are stuck in the intracellular fluid.
In response to your what if statement, I think that each species would have a different limit that tells the organism to either cheat or produce. This would require more research and would be difficult to control because of all of the different interactions present because basically, ecology. I honestly don’t know how an experiment of this type could be set up. Definitely an interesting thought experiment.
The best way that I am able to wrap my head around this section is that vF is trying to monopolize its niche so that vH is unable to displace it. Earlier in the paper it was shown that vH usually outcompetes vF when in a richer media.
The organisms that consume the oxygen are engaging in aerobic respiration and were not the object of the study. I would say that the organisms responsible for the generation of the hydrogen sulfide are the decomposers at the seafloor and also the hydrothermal vents that release a lot of hydrogen sulfide.
I was wondering what natural resources are specifically provided by Antartica. Due to my curiosity, I googled “what natural resources does Antartica have?” I discovered that Antartica has: antimony, chromium, copper, gold, lead, molybdenum, tin, uranium, and zincs. The paper states, “oil spillage due to transport, storage and utilization of fossil fuels has become a serious and persistent threat.” When searching for my previous question, I found out that some people have considered towing icebergs from Antartica in order to provide fresh water to other parts of the world that are in need. If this was to be done, would it do more harm than good?
The paper explains how oil contamination can generate detrimental changes in soil properties. From that, physical and chemical changes promote rearrangements in the soil bacterial communities. This is causing a huge decrease in species richness and soil biodiversity. Oil is a natural resource that we use in everyday life, so I do not think we are going to stop using it anytime soon. With that being said, is there anything we can do to limit the amount of contamination and/or decrease the effect it has on the ecosystem?
The paper states that they used R2A agar when plating the samples. What would the results look like compared to the same samples on TSA, PEA, or McConkey?
What does it mean when it says “cultures were grown with agitation”? Does the excitement of the agitation cause them to grow more rapidly? What is the agitation exactly?
While looking at Figure 4, I noticed that E.coli BW25113 and P. guineae E43FB had low data points in B. However, in A P. guineae E43FB performs higher while E. coli BW25113 is still near the bottom of the chart. What is causing this change to occur?
I’m sorry. I meant Figure 2!
The paper states that the D43FB strain from Antartica exhibited the highest growth yield. If those same three strains were taken from a different area, such as the Tropics, would D43FB still be the highest yield?
Hey Jacob! I read your comment and thought the same thing, but then I had to know ‘how bacteria do perform in environments like Antartica?’ With that being said I did some googling and discovered that extremophiles known as psychrophiles are known for surviving in cold temperatures. Different species adapt in various ways. However, most trigger a physiological response that allows them to adapt to these unfavorable conditions by making changes in membrane composition. The microorganisms will also adjust translation and transcription machineries. The response from the cold prompts a growth block, as well as repression of translation. Along with this, a set of specific proteins are introduced to make sure the cell metabolism is back in tune and readjust to the new conditions. It also found out that this adaption for E. coli would take about 4 hours.
I know this does not answer your question about the diversity (I am not too sure either) but maybe this is one reason that there can be bacteria diversity in such harsh condition environments like Antartica?
[Microbial bioremediation is quickly becoming an important approach in the continuing efforts to decontaminate critical sites of oil spillage in cold weather environments]
This statement caught my attention because of the “in cold weather environments.” Does this mean that Microbial bioremediation is solely done in cold weather environments, or does it just perform exceedingly better in these conditions compared to somewhere with warmer temperatures? I did some googling and learned that oil pollution in harshly cold weather has led to a higher vulnerability towards the petroleum pollutants compared to tropical and temperate environments. Although I am pleased to acquire this information, my question above still stands. I also apologize if my comment did not clearly reflect my thought process. Basically, I just want to know- is Microbial bioremediation beneficial for other environments?
I am confused with this paragraph. In the beginning it says, “Iron is an essential nutrient for virtually all organisms,” but then states, “iron acquisition presents a challenge to most bacteria.” So, is iron benefitting or hindering bacteria? Does it depend on the situation?
I was interested in the Vibrio species. After I googled it, I found out the siderophores (molecule that binds iron) from these species actually developed through evolution. Since this is a developed system, what did the species do before then to acquire iron?
It never crossed my mind to use iron depletion as a source to induce competition. What other ways are there to promote competition when observing growth in bacteria? How different would this be compared to an environment with an excessive amount of iron?
The scientists screened the colonies for chloramphenicol sensitivity. I did not know what “chloramphenicol” was and had to google it. I found that it is used as an antibiotic for many bacterial infections. It is found in eye ointment and also treats plague and typhoid fever. I think it is neat that chloramphenicol can be used in everyday life products and to help with DNA manipulation and mutant construction!
I wasn’t 100% sure what “arbitrarily-primed PCR” was, so I googled it and discovered that it is a DNA fingerprinting technique- so cool!
It says “crenarchaeal amoA activates in the Black Sea water column.” Is that the only place that it activates?
[For decades, only specific groups of β- and γ-proteobacteria have been found to exhibit this capability. However, recent metagenomic studies in the Sargasso Sea (16, 17) and later of a marine sponge symbiont (18) have identified in marine group I (MGI) Crenarchaeota genes encoding proteins resembling ammonia monooxygenase (AMO), the key enzyme in aerobic ammonia-oxidizing bacteria (AOB).]
Only specific groups were able to exhibit this capability until recent studies. Was this a completely new discovery or did advances in technology help us find this?
I wasn’t sure what “flow cytometry” was, so I googled it. I found that it is a technique used to detect and measure physical and chemical characteristics of a population of cells or particles. It is measuring the volume of cells in a rapidly flowing fluid stream as they pass in front of a viewing aperture.
It is mentioned that gas chromatography was used. This type of chromatography is the process of separating compounds in a mixture by injecting a gas or liquid sample into a mobile phase. It is typically used to test for purity or to separate components of a mixture.
Zachery,
I was looking at the figures and was trying to figure out how to put into words what I thought. The way you expressed this was great on what I was trying to convey!
CARD-FISH is something that is new to me, so I wanted to learn what it was. I found that it is an updated version of the traditional FISH. It is used for phylogenetic staining of microorganisms in various environments. The use of this has helped elucidate the microbial ecology of many habitats.
In Black Sea suboxic waters, microaerobic nitrification is directly coupled to anammox. Would this be the same for different water areas?
If Crenarchaeal amoA mRNA variation can explain 74.5% of the nitrite variation within the oxic zone, and AOB amoA mRNA only accounts for 6.5%, what should we expect to be the cause of the remaining 19%?
If bacterial biodegradation is the major contributing factor to cleaning PAH contaminated ecosystems, and PAH’s serve as growth substrates for bacteria; why is bacterial degradation only able to metabolize lower molecular weights? if the bacteria grows in response to PAH’s feeding should it not be able to phagocytize PAH’s of larger molecular weights?
Cyclic aromatic compounds are stated to to afflict great harm to animals and a toxic mutagenic compound which can cause cancer. if these toxins multiply at a very high rate how will bioremediation on its own filter out the toxins? When it is stated in paragraph three line six that bacterial biodegradation is not characterized for high molecular weighted PAH’s ?
The concentration of the turbidity of the water samples obtained to measure the effects of Naphthalene degrading bacteria would be measured by an Assay to analyze the replication of DNA.
what made up the culture medium broth? why was a flame ionization detector necessary for the purpose of agitating the vortex by centrifugation?
what is the significance of using a neighbor joining analysis ?
what is the GC-FID method?
gram positive bacteria have a thicker cell wall and can handle the high variations of environmental stress unlike gram negative bacteria. and this can be a factor as to why gram positive bacteria plays an important role in naphthalene degradation.
the screening of naphthalene bacteria causes high diversity, but how is there a high diversity of the bacteria destroys its like?
what does it mean when a cloned gene is partially characterized?
what characteristics of grouping from the phylogenetic tree did you look for upon choosing the type of strain?
What was the purpose of adding the Bushnell-Haas minimal salt medium to the soil samples? what effects does it have?
What were some of the characteristics to colony morphology that you take into consideration upon choosing colonies to isolate. i want to pick up this technique to help me with spotting good colonies in the lab as well.
how does the flame ionization detector help gas chromatography? does it measure the amount of oxygen being used or CO2 being produced
it was noted int his paragraph that there were physical color changes in in the sample of bacteria, what were the noted color changes and was it considered positive or negative?
How long was the incubation period for the BH liquid cultures, if it was the classical 24-48 hours such as in lab, and there were still no signs of growth then what precautions would you have taken to stimulate the growth of the bacteria. i know in the lab the zinc was added for the nitrate reduction to help the process of negative response.
what are the four main clusters that were formed int he phylogenetic tree?
was the appearance of a blue indole color a positive or negative indicator, and why?
Open reading frames (ORF) is found in bacteria or archaea genomes. a functional ORF can encode a protein electronically using plasmid pUC119. the process of finding and identifying ORFs begins by the computer finding the start codons and any possible stop codons. the codons are then processed by finding the codon count and possible ribose binding sites. finally the ORFs are organized in a list.
Based on my previous comment the indigo color indicates a gram negative bacteria. from my understanding a Biolog-GN test is a gram negative test is for the purpose of identifying aerobic gram negative bacteria by analyzing the metabolic fingerprint . did you choose the biolog-GN method for fingerprinting the bacteria because it works alongside gram negative results and has a very large number of tests?
Bacteria GST stands for glutathione transferases, in which is an enzyme that participates in detoxification as an aerobic bacteria and is a good method for observing the binding of proteins in an in vitro scenario.
after the oxic and anoxic bacteria has con through incubation under conditions with and without oxygen, afterwards an aerobic and anaerobic oxidizer were each individually added as a defined co culture. oxygen tension meaning different concentrations of oxygenic conditions were used to test its relevance to the bacteria.
the extreme conditions of a hot spring and the gases might closely mimic the environment of early earth an anaerobic conditions, the purpose of this lab might be to find out how the organisms of early earth survived under the extreme conditions
it says that the red colored biofilms were collected through out a large period of time with long gaps in between: August, October, and April. do the climate change in the seasons of these months that the samples were collected not have a variable in the experiment?
i thought the purpose of using N2 gas was to create an atmosphere without oxygen, to resemble the atmosphere of early earth, was the foam stopper the next step to allow gas exchange in small amount to resemble the introduction of oxygen into the atmosphere like that of early earth?
looking at figure 4 , if AsV is reduced is it an electron acceptor because it is gaining electrons
Figure three shows that AsIII is the electron donor and AsV is the electron acceptor. the graphshows a comparison in the temperature ranges Vs. the oxidation of AsIII and the reduction of AsV: AsIII shows a peak at 43 degrees C with a broader pemperature range, and the reduction of AsV shows a narrower temperature range but a comparable optimum.
methanogenisis is a process that is very important to anaerobic breakdown of organic materials in the absence of oxygen to allow the formation of a gas mainly composed of carbon dioxide and methane.
AsIII has a broader temperature range due to the oxidation rate occuring in the dark with no light driven photosynthesis and AsIII is an electron donor in this reaction and a chemolithtroph.
if the springs and the ponds were less shallow and much deeper would the amount of maltonic microbes be fewer and in turn there would be no detection of of aerobic microbes due to the environmental conditions? in this case would the values of AsIII oxidation in figure three be even broader with no peak in temperature optimum?
chemoautotrophy stimulated the reduction of As V
Why in particular is Antarctica a unique territory in earth’s climate as well as a source of natural resources? It is due to the ecology of the location, climate, natural occurring microbes, etc.
“Oil contamination can generate detrimental changes in soil properties.” Does the rearrangement in the soil bacterial contribute to the growing crisis of climate change which can be seen by the increase in surface temperature, pH, and carbon and nitrogen levels?
To my knowledge, after Googling, 16srRNA has to do with identifying/differentiating between bacteria species. Were certain species of the 350 bacterial colonies selected more prominent than others? Also, did the of acquiring diesel/non-diesel fuel colonies have an impact on which colonies might have an increase in the metabolizing of phenanthrene and were these bacteria related?
Since using a Scanning Electron Microscope is a great way to view biofilms, where their certain colonies out of the diesel/non-diesel fuel which yielded a greater cell adhesion to the phenanthrene? Of these colonies, were there any which made up a greater portion of the biofilm in the “Biofilm Formation Assays”?
“In contrast phenanthrene supplementation of diesel-exposed soil promotes sudden bacterial growth after 72 h, suggesting that phenanthrene is being metabolized and used as a carbon source by the bacteria present in contaminated soils.” What about the chemical structure of diesel-exposed soil allowed the bacteria to utilize the phenanthrene? I tried to google the chemical structure of diesel, and all I found is that it consists of 25% aromatic hydrocarbons, does this play a part in the metabolism of phenanthrene?
“The phenotypic characterization showed positive utilization of substrate for the three stains for D-Glucose, L-arabinose and D-maltose.” Why would the strains not show characterization of L amino acids since that is normally the preferred configuration? Also, if you were to switch the substrates for D-L-D to L-D-L, would the results then show negative utilization of substrate?
It fascinates me that isolate S. xenophagum D43FB not only showed remarkable degradation, but also managed to follow kinetics of growth with other isolates by differing research groups. I would love to know if there are any other regions, beside Antarctica, where this isolate inhabits and would it degrade/replicate at similar rates as it does in Antarctica.
My assumption was the SSMS approach was used to help identify the most potent PAH metabolizers. I do agree with you on “Xenobiotics need to be studied more as a whole.” This was my first time hearing of the term while reading this article. After doing some quick Googling, it astounds me how much impact they have on environmental/human health.
So I googled “direct competition strategies” and found it occurs when individuals are competing for the same resource. If that is the case, are microbes able to utilize transformation of dead microbes DNA in order to gain antibiotic resistance those same microbes exhibit (i.e. production of antibiotics)?
I’m confused by the following “When two or more species each produce siderophores, the species that produces the siderophore with the highest affinity for iron can enjoy a competitive advantage.” Would all siderophore producing microbes not have the same affinity to iron? If not, I guess I’m curious as to what impacts these siderophores ability to “bind” to iron?
“Unless otherwise indicated, erythromycin, chloramphenicol, kanamycin, ampicillin, and polymyxin B were added to final concentrations…” I am confused why the selection of these antibiotics in specific were chosen. Do these play a larger impact when dealing with plasmid introduction?
So I googled Oligonucleotides and found these are short DNA/RNA molecules which have a wide range of applications for testing. I am assuming oligonucleotides are to be used for the “priming” process of PCR. Is there a specific reason oligonucleotides have to be the priming source for PCR?
“We conclude that iron chelation is responsible for the inhibition of V. harveyi growth when V. fischeri ES114 culture fluids are present.” I had to google “iron chelation” and found it is the removal of excess iron through the introduction of special drugs. With this, why would culture fluids from V. fischeri ES114 allow iron chelation to inhibit growth of V. harveyi?
“We conclude that IutA and FhuCDB, encoding, respectively, the siderophore OM receptor and siderophore importer are sufficient to convert a non-aerobactin-producing species into an aerobactin cheater.” This helps answer my question as to “How” non-aerobactin are converted into aerobactin cheaters. However, is there any particular use to being converted to a “cheater” as compared to someone who is already able to utilize iron?
So I had to look up “upwelling regions,” and I found it is when nitrogen-enriched, deep water is moved towards the surface. Is denitrification/anammox more likely to occur at surface level regions as compared to deep level regions due to specific microbes that aid in the nitrification process?
“The first and rate-limiting step of nitrification is aerobic ammonia oxidation.” I am a little confused by this. Since the process is aerobic, it requires oxygen for ammonia oxidation. Yet the Black Sea has low levels of oxygen (I’m assuming the deeper you get into the sea the less oxygen is present) so how are these microbes able to nitrify? Is this why MGI Crenarchaeota is an important nitrifier?
It is interesting to see the abundance of species that are present at 80 m as compared to the other two regions. It appears that as the depth increases, species expressed decreases. I also see BS160B4 is able to be expressed at 80 m, 100 m, and 110 m.
It appears mRNA is able to thrive in suboxic levels in all figures specifically that of Figure C and D. I notice in Figure A, the cells are increasing as mRNA is decreased between 100 m and 110 m.
“It is possible that V. Fischeri ES114 switches between siderophore-producing and siderophore-cheating based on iron availability and whether or not other vibrios are present that can supply siderophores.” The idea of a species being able to “switch” certain characteristics is always fascinating to me. Curiosity just makes me wonder how this species was able to accumulate both producing/cheating traits over time.
“While highly speculative, we suspect that glpK mutation demands that V. Fischeri ES114 use amino acids, rather than glycerol, for growth, altering metabolic pathways that decrease the availability of a substrate required for aerobactin production.” I am a little confused by this statement. What specifically about glpK mutation causes V. Fischeri to utilize amino acids? Also, would this be a beneficial mutation since it “decreases the availability of a substrate required for aerobactin production”?
“Sulfide concentration was measured onboard spectrophotometrically (47).” I remember reading about some bacteria in marine sediments which can transfer the electrons from sulfides and deliver via “chains” to other areas where oxygen is present. I wonder if this is the case here. Maybe the microbes are transferring electrons from oxygen deprived areas to better oxygenated areas of the water? Just a thought.
Brianna, I had the exact same thought. After Googling “Steady State Flux,” it appears to be a constant concentration gradient like you stated. I’m now wondering if this steady state is being used to assist in processes of nitrification.
“Despite the barely detectable gene abundance, strong amoA expression by AOB was detected within the nitrification zone (Fig. 2).” Based on the graph from the previous discussion, this makes sense; however, I wonder if strong amoA expression would be observed in other seas around the world when compared to the Black Sea?
Makayla, I had the same question. I agree it probably has to due with nutrient availability as well as oxygen, pressure, and light available at these depths. I would honestly love to know more about BS160B4. The fact it was found in all three depths was astonishing. I also wonder if it is found else in oceans and exhibit the same characteristics?
If bioremediation is, in part, dependent on the molecular weight of PAHs, would the catabolic activity of biomolecules be hindered or promoted due to a pH change in the environment?
Would it be possible to “condition” naphthalene degrading bacteria in order to allow for habitation of multiple otherwise inhospitable environments? Maybe via selective breeding?
Storing the growth media at -20°C inhibits the growth. What is the reasoning behind not wanting the media to continue to grow?
Is knowing the evolutionary history a valid way to characterize naphthalene degrading bacteria because of the many possible phylogenetic outcomes?
There seems to be a positive correlation between growth and the percentage of naphthalene degradation. This is not a linear trend, because some strains with lower growth have a higher percentage of naphthalene degradation. Could this be due to some strains having a lower volume to area ratio, therefore their growth increases effectiveness?
Once concentrations of naphthalene get above 400 ppm, are the naphthalene-degrading bacteria not able to degrade due to their lack of size?
Were all 54 naphthalene-degrading bacterial strains used in experiment, or is it just worth noting the variation?
What uses could narrowing down the most ideal naphthalene-degrading strands bring?
Because bacteria Cycloclasticus are limited in usability of carbon sources, would they still be the ideal bacteria for aromatic degradation or is it not difficult to keep them alive?
The ability of marine bacteria to degrade more readily could be due to a higher abundance of nutrients for the PAH degrading bacteria to obtain carbon from.
Using both mass spectroscopy and NMR allow identification of the molecule’s molecular mass, which gives insight to stability and reactivity of the molecules. Also NMR gives information regarding the movement of the molecule’s atoms, further allowing for properties of the substance to be interpreted.
Why is ORF7 reading in the reverse direction? Is this the one partial ORF that was found?
Do these other Beta subunits not show much similarity because they were not extracted from PAH dioxygenase?
Knowing this dominating bacteria are alongside halobacteriacaea indicates the ability to absorb light and use for ATP, but can also function as aerobes.
Arsenic can be utilized in redox reactions under anoxic conditions if there is light present.
If the groups did not produce two or more clones were they not considered viable?
Why was this bacteria not tested in a usually hospitable water solution that contained arsenic? That could provide more insight to amounts of arsenic/liter this bacteria could reduce.
They probably used RFLP analysis in order to characterize these bacteria because they were found in a mixed-species community.
What occurs between the times of 21 hours and 27 hours that changes the speed at which electrons are donated so drastically?
Would the ability for arsenide to oxidize without an electron donor present indicate the presence of toxins due to long incubation time?
Their main purpose of this paper was to determine the certain microbe’s use of arsenic in the environment in Mono Lake in order to study HOW these microbes interact with arsenic. The information gathered from this experiment could be used to study and compare the activity in different environments.
Because the Archaea found in this same system are not involved in arsenic cycling, the Ectothiorhodospira are independent when dealing with redox reactions of arsenic and can probably be useful when isolated and placed in a different environment.
Would an environment which supported oxidase genes improve axoB activity? Is there a reason for not testing in an environment which supports oxidase genes?
Why would they not study the anaerobic pathway since the bacterium was found in deep sea sediment?
The differing oxygen levels in marine vs terrestrial environments would play a large role in differences in anaerobic and aerobic pathways in the bacteria, producing different results.
What is the importance in generating a gene deletion mutant? And would this be a multi gene or single gene deletion?
If the strain can grow with fluoranthee as the sole source of carbon and energy, it is able to degrade the compound. What metabolites would researchers be looking for in this situation?
Were these nine genes expected to encode different ring-cleaving dioxygenases based off of previous research or based off of their similarity to one another?
What is the purpose for studying strain B30? Is it strictly for comparison to deep-sea sediment or based off of its likeliness to Celeribacter baekdonensis?
“However, the taxonomic distributions of the five plasmid proteomes of strain P73Twere different from that of the chromosome, suggesting the chromosome and the plasmids may have had potentially different origins.”
This could be due to horizontal (lateral) gene transfer which led to a divergence in its evolutionary history.
Lateral gene transfer is important in this study because the new genetic information introduced into the P73T strain could be unique to its species.
How would the P73T strain survive environmental conditions with only one viable deoxygenation pathway?
Finding out these physiological features of the bacterium would allow researchers to gain a better understanding of how it could be used in real world settings.
Is the identification of the enzymes that govern the initial attack necessary in identifying PAH degrading bacteria, or would it be helpful in the bioremediation using this bacteria later on?
Would it be necessary to clone these known genes and introduce them to a host that would tolerate fluorene as a sole carbon source?
What is the importance for such a rapid and drastic increase in incubation temperature? Is it necessary because of the presence of helium gas?
Could the silicone oil be further tested (through Infrared Spectrometry or Nuclear Magnetic Resonance) to determine which PAHs were degraded by the bacterium?
So there is presence of recombinant proteins but they are not active, does that infer the recombinant DNA encoding for the protein was never introduced into the host cell? If so why is this information significant?
Fluoranthene, dibenzofuran, and dibenzo-p-dioxin all yield the highest % product, so these are the best substrates found in the table. Fluorene has a low % yield and produces dihydroxyfluorene, 1-hydro-1, 1a dihydroxy-9-fluorenoned, and fluorene-dihydrodiol. We see that dihydroxyfluorene and 1-hydro-1, 1a dihydroxy-9-fluorenoned are not present in DFDO.
The sphingomonads’ relationship to other pseudomonas is interesting considering it’s more relevant in human diseases rather than wastewater-type treatment of PAHs.
Has any other research been done to identify other bacteria that degrade these PAHs?
Why is the donor-recipient ratio at 1:1? Would a more skewed ratio impact the amount of product produced or their quality?
Do these strains just take longer to be cultured than most? Or are all of these incubation processes done for another reason?
Does this 40% homology give us any valuable information, or is it too low of a commonality to make a difference?
What is the purpose of converting these molecules to their dihydrodiol products? Does it help in identification or methods involved with metabolism?
Wouldn’t the inability to oxidize indole to indigo be an unexpected result? And what element of the DNA was unstable? I feel like in the results section that should be classified and then further explained in the discussion.
Strain A4 serving as a substrate assists in the strain’s ability to degrade these PAHs, but was work done to see if they were degraded without the presence of this substrate?
I think finding its homologs would be a great place to start, and then further the research by testing the strain’s ability to degrade these PAHs in an environmental setting.
I believe the purpose of this research was to set up a set of information necessary to carry out further research into the degradation of these PAHs. Other researchers may be able to use these now known PAH-contaminated sites and construct gene probes specific to the sphigomonads in order to leran more about their PAH-dioxygenases.
Do these gene clusters communicate with each other by folding? Otherwise I do not understand how these gene clusters can work together to serve a similar function even with the same sequences.
Glycosphingolipids are involved with signaling between cells. These lipids could be involved with the separate gene clusters working together to serve a function.
This also allows researchers to isolate the gene of interest when comparing it to the original sequence
When would it be necessary to utilize ampicillin, chloramphenicol, kanamycin and tetracycline?
How can you tell when the strain is in the stationary phase? What happens if the strain is left in LB medium for too long?
What is the benefit of inserting the GMr cassette into the disrupted regions?
Does extending the primer allow for a wider region to be found in PCR?
The loss of ability to produce indigo shows that the genes which were disrupted probably contain RHD
ORF 11 being a psuedogene, would there be any further research to determine a possible function of this ORF in a gene that has not been disrupted? Or is it produced solely because of this experiment?
ORF6 seems to put in more energy to run transcription than it gets out from transcribing, therefore proving it inefficient.
Is this to say ArhR overproduces mRNA regularly unless an induced is present?
The article cites dermal and respiratory uptake and direct or indirect ingestion as potential ways we can become exposed to PAHs. It goes on to say that prolonged exposure to PAHs can result in cancer of the lungs, heart, or kidneys. This particular fact caused me to broaden my thinking past PAHs to consider what other toxic compounds we can unknowingly come in contact with during our day-to-day lives that put our health at risk.
Jennifer, I too was curious about what caused the change in homology. The ultimate source of evolution is mutation, so I wonder what things in the Rodococci’s environment added up to have an evolutionary effect on the microorganism. With metabolism, growth, and evolution being a universal trend in all organisms, I believe considering the cause of change in Rhodococcus sp. CMGCZ’s homology to be a very appropriate inquiry.
The vast amount of media options available in microbiology are very intriguing to me, but almost quite daunting. I’d be interested to know what steps a microbiologist takes to analyze the different media options in relation to their experiment in order to optimize growth conditions and provide valuable results. Furthermore, does media choice greatly change results, and how do you know if experiment failure/success is a result of media choice or something else?
The paper cites Nap, Phe, and Fla as being the sole carbon and energy sources for this portion of the experiment. What is the purpose of ensuring that the PAHs are the sole carbon and energy source? Is it to ensure that the bacteria are going to seek these out for metabolism and subsequent degradation?
I find it very interesting that the Fluoranthene was completely degraded after one week of incubation. What contributes to my interest in this figure is that none of the others were completely degraded, not even close! I wonder why Fluoranthene was “easier” to degrade than Napthalene and Phenanthrene; I would’ve assumed a low molecular weight PAH, such as Napthalene, would have degraded quicker and more thoroughly than a high molecular weight PAH such as Fluoranthene.
I’m interested in the relationship between time, degradation, and medium choice. More specifically, the fact that the MSM allowed for quicker degradation at the beginning, as opposed to the YMSM which had slow degradation at first followed by a rapid phase of degradation on the 5th, 6th, and final days of incubation. What benefits to bacteria growth/effects on degradation rate does minimal salts medium provide over yeast enriched medium, and vice versa?
I find it interesting that Rhodococcus favors Fla so heavily. Fla is a four membered ring, or in otherwords a High Molecular Weight PAH. With napthalene being a Low Molecular Weight PAH with only 2 rings, I’m surprised that the results supported the conclusion that Rhodococcus sp CMGCZ prefers a HMW PAH, such as Fla., over a lower one, such as nap.. I wonder what contributes to this preference – more carbon, chemical stability of the differing PAHs, etc?
The paragraph states that the Rhodococcus sp. CMGCZ was enriched on Fla in its original growth, and after which was found to degrade 99.3% Fla. It goes on to say that after being continuously subcultured, strains were isolated that were able to grow on Phe. I find it so intriguing how adaptive the bacterium were in order to be able to survive on Phe instead of their usual Fla.!
It seems to me that an organism would prefer to metabolize a more easily degradable carbon source, such as YE, over a difficult one, such as a PAH. If the goal is to use as little energy as possible to carry out the processes necessary to maintain life, then YE seems the way to go over the polycyclic aromatic hydrocarbons. So why then would the researchers in this study choose to provide the bacterium with an “easy way out”?
I’m happy that the paper informed us of how the information gathered from the research would be used. I’m interested to learn more about what specific scenarios and locations these bacteria are used to break down PAHs. Must they be added to an environment, or is the naturally occurring PAH degrading bacteria enough to handle the biodegradation, etc. efforts on their own?
Jennifer, if I remember correctly, the first paper we read discussed a few ways that humans may come in contact with PAHs, one of which being ingestion. This paper cites seafood consumption as a means of ingestion, while the other states that PAHs can be ingested by humans directly or through the food chain. The other ways mentioned in paper one include dermal and respiratory uptake.
Nolan, what an appropriate and necessary thought process! We as consumers need to be made fully aware of what we are ingesting, and what benefits and/or consequences the food in question may bring. I definitely would be interested in learning more about any correlation between PAHs in food and occurrences of cancer. I too would like to know what precautions are taken by food providers.
I am looking forward to reading more about the PAH degradation enabling enzymes possessed by Cycloclasticus sp. strain A5. I hope to be able to compare and contrast the degradation abilities of this marine bacterium and the Rhodococci discussed in the previous paper. I wonder if there is a parallel between the enzymes of the marine bacterium and the ARHDs of Rhodococcus sp. CMGCZ.
I am curious to know the evolutionary history of the genus Cycloclasticus. What environmental conditions and or genetic mutations initially allowed it to degrade aromatic compounds?
Jennifer, I too think it is so cool that we are getting to put into practice what we are reading about in these papers. For this reason, I am very much looking forward to this week’s lab when we get to isolate our bacteria’s DNA! Furthermore, we will get to use the DNA to make phylogenetic connections for our bacterium based on the family of enzymes mentioned in the papers.
I have two questions regarding this portion of the paper:
1. What was the purpose of supplementing with antibiotics?
2. What is the purpose of expressing the gene in a different species of bacteria? Furthermore, why was the lac operon of interest for the study?
I think it is so cool that we are able to determine metabolic properties of microorganisms just by looking at their sequencing. I find it even more interesting that in this study, the phn genes were not clustered as they were in previously reported aromatic ring dioxygenase genes. I wonder how this sets Cycoclasticus sp. strain A5 apart in terms of both evolutionary history and PAH degradation potential. I really look forward to the paper hopefully answering these questions!
I’m intrigued by the phnC gene, as it comes from a subfamily with such diverse sequences. This diversity makes me wonder what happened from an evolutionary perspective to result in such a numerous amount of similar but different genes in different bacterial species. Furthermore, the fact that the enzymes of this group show high activity with different substrates seems promising for bioremediation efforts. Notable aspects of A5’s family, in relation to bioremediation, is their ability to cleave bicyclic substrates.
Madeline, I too was interested by the paper’s mentioning of this difference. It’s strange that A5 differs in such a way from what is “normal” of PAH degrading bacteria. I wonder if the fact that the genes are located on a chromosome instead of a plasmid plays a role in A5’s wide substrate range.
Lane, these evolutionary questions are precisely what I was considering upon reading this section of the paper. The paper cites that the A5 strain has many substrates, and I’m sure this has a lot to do with the overall complexity of the gene cluster we are reading about. Furthermore, I wonder what environmental stressors this bacteria strain endured to have to adapt to multiple substrates in such a way.
It will be interesting to see if and how they experiment with oxic vs. anoxic conditions. After our POGIL in lab today, I am curious as to if this would be a good situation to utilize transcriptomics to look at and compare which genes are active under either condition.
I’m looking forward to reading more about the conclusions drawn from their research. The discussion regarding the role of these metabolic pathways in the Archean Earth is intriguing!
I’m curious about the evolution behind these bacteria’s ability to use arsenic metabolically. Has the species of interest and its ancestors always been able to do this, or was it an evolutionarily advantageous adaptation?
I too thought it was very interesting to think back on our previous papers’ bacteria in comparison to this one. In the previous papers, the research has been utilized for bioremediation purposes. I am curious as to if these bacteria are capable of similar benefits.
I wonder what role sulfide, ammonia, and methane could play for these bacteria. If the arsenite is unavailable which is the most logical next choice? Are these bacteria able to metabolize such properties? I look forward to diving deeper in to metabolism and redox potentials so that I may answer this question myself!
I’ll be interested to see any phylogenetic trees based around these genes presented by the researchers in the results. I’m also interested in seeing if these oxidative genes bear any structural resemblance to those of the previous papers? I know this paper is discussing the metabolism of arsenic and not PAHs, but still.
The paper states that the oxidation rate lowers after each subsequent reimposition of light. Does this occur because metabolic pathways that are more efficient in the dark are being selected for during that time without light, and then out competing others so that when the light is reimposed the bugs present are less efficient at oxidizing AS(III) to As(V)?
In comparing figure 1, anaerobic As(III) oxidation and As(V) reduction, to figure 2, aerobic As(III) oxidation and As(V) reduction, it appears that the reactions are occurring faster under aerobic conditions. I predict this is because the energy generated from aerobic respiration is much higher than that of anaerobic, thereby speeding up the reaction times.
It’s interesting to think about how senescene over time affects metabolic processes across the domains of life. As seen here it dampens the rate of cycling, but this reminded me of how biological aging and mutations affect processes in the human body.
The paper states that different microbial populations play a role in the observable reactions.I think further experimentation and analyzation using metagenomic techniques would be interesting here.
Cyclic aromatic compounds are mentioned as causing significant harm to mammals, however no statistics are given along with this citation. I am interested to know exactly how often or what percentage of the population suffer from exposure to these substances each year. Also, what countries or communites are most impacted by these substances?
Have these microorganisms been isolated in our area and observed in relation to the 2010 Deepwater Horizon oil spill? If not, could this strategy, if effective, also be used to help our own community?
The materials and methods fails to mention how many samples of the bacteria were cultured. Culturing more than one would certainly be important in order to ensure that no contamination occurred and consistent results were obtained.
What exactly is causing the changes to cell surface hydrophobicity and how does it relate to the overall purpose of the experiment?
Is the decrease in naphthalene degradation after 400 ppm a result of sensory overload? If after 400 ppm the bacteria are unable to grow as much then would they eventually be killed by a sufficientally large naphthalene concentration?
I would be interested to see a study that followed the growth rate of these bacteria on a daily basis and in the presence of different concentrations of naphthalene. Would the rate of their growth increase or decrease over time and how would each strains rates differ?
Have more studies been done on this subject at the Persian Gulf since this one was published? If so, are the results consistent and were any gram-positive bacteria ever isolated?
In relation to a previous comment I made, if naphthalene degrading bacteria are present on our own gulf coast in relation to the Deepwater Horizon oil spill, then I wonder if they would be gram-positive or gram-negative. I think that could be a very interesting study to perform with it being so close to home for us.
Bushnell-Haas minimal salt medium is used to determine the ability or inability of an organism to degrade hydrocarbons. This is important for this experiment because it is looking at the degradation of the hydrocarbon phenanthrene.
I looked up and found that sonication is the disrupting of cells using high pitched sound waves. However, I also saw that it is very expensive and usually inefficient. If that is the case, I am curious to know why they chose this method. Was it the only method available for what they wanted to achieve?
Are phthalic acid and diphenylamine unable to support the growth of ZX4 because of their different structures? In other words, the structures are quite different than those of phenanthrene, fluorene, and naphthalene, so is that why they are not useful?
I would think that the lower C230 activity when strain ZX4 is grown only with glucose coupled with the 98.74% of phenanthrene being degraded, indicates that phenanthrene is a better carbon source for it. Is this correct? If so, then perhaps this strain is a good organism to use for bioremediation, assuming it doesn’t have any harmful factors that have not been indicated.
What exactly does the number of base pairs in between the phnH, phnG, and phnI mean? It is also very specific about the number of base pairs upstream from the start codons. What is significant about this fact?
When I looked up Sphingomonas paucimobilis, I learned that it actually used to be a part of the species Pseudomonas. I know these are bacteria that can cause infections in humans. As I search further, I found a paper that looked at the infections caused by this particular strain in patients. Since it is potentially harmful to other organisms, particularly humans, I wonder if the authors of this paper were aware of that when determining it was a good candidate for bioremediation. Perhaps they did, but the strain doesn’t cause enough infections to matter in the long run?
I think I may be getting confused with all the words here, but is this paragraph just saying that the arrangement of these genes is what helped to determine the bacterium the strain was based on the arrangement of the meta-pathway genes?
If I understand correctly, it seems like they are implying that perhaps the genus Sphingomonas may have risen from horizontal gene transfer from the already established Pseudomonas genus. Is this correct?
I looked to see what other toxic elements some bacteria and archaea may be resistant to, and they included bismuth, cadmium, mercury, and several others. I wonder why the authors chose to research arsenic as opposed to one of the other possibilities. Perhaps it was due to ease of experimentation?
When they say “uncontaminated soils” do they mean that the soil is not contaminated with arsenic? If that is the case, then do they offer any theories as to how the organisms developed the ability to reduce or oxidize it?
I don’t really understand what it means when they say they subsampled the slurries each hour with an N2-flushed syringe. Does this mean that a small amount was taken out and reinfected into a new incubation setup?
What purpose does knowing the residual radioactivity serve for this experiment? Is it in order to see the dangers that are still being exposed by the organism even after death?
Were the archaea findings more diverse because they are able to survive more easily in the environmental conditions?
The way that the organisms are able to switch back and forth between As(III) and As(V) leads me to believe that the pathways and the needs of the organism are being controlled by the presence or absence of light. The figures seem to show that when light is present, the As(III) is being oxidized to As(V), and when light is absent the As(V) is reduced back into As(III).
Could the researchers perhaps take this information and and conducts a more pointed experiment that focuses on just bicarbonate assimilation using more than the two they did here? I just wonder if there may be one that is even better than these two when in the presence of hydrogen and sulfide.
I would like to see research done to isolate even more microbes with these capabilities, and determine if these genes are also contributing factors.
Does this mean that they do not think that Ectothiorhodospira contributed to the dark reaction data at all? If that is the case, then what strains are causing such a high temperature range and rates?
Could we not study the properties of the PAH-metabolism process and form a synthetic version of it for mass use and production? I can see that that would be easier said than done, but it may be a good endpoint to strive toward.
Why exactly do they feel that this particular strain will be a good model? What specific qualities are leading them toward that conclusion?
Reading the effects oil can cause on the bacteria living in the soil, remembering the oil spill… Makes me sick. My daughter caught pseudomonas when she was 2 and she will likely have it her whole life, unless of course she receives new FDA approved treatments when she turns of age. Interesting to read that “specially Pseudomonas – dominates over Bacteria that compose the normal inhabitants non-polluted soil.”
I found another similar article discussing King George Island. Doing so I discovered, Different studies on the contaminated soils have revealed the metal is directly interacting with the microbial community. Antartica uses Diesel Fuel as their main source of energy as well as contains 80% of the freshwater reserves of our planet.
Gran-Scheuch, A. (2020, November 7). Effect of Co-contamination by PAHs and Heavy Metals on Bacterial Communities of Diesel Contaminated Soils of South Shetland Islands, Antarctica [Editorial]. Microorganisms.
I had to read more about the KEGG database. I searched and found the database overview which informs readers that it is used as a reference knowledge base for interpretation of large-scale data sets generated by genome sequencing. I also learned, KEGG is being expanded towards integrating human diseases, drugs, and other health related substances. Last updated November 1, 2020
Why not include samples deeper than the surface soil? I would suggest the diesel fuel would absorb into the surface and reach even stronger concentrations deeper into the soil. Transported on ice, makes me wonder if any foreign temperature change would either grow or kill the bacteria.
Do oil fields apply to places where oil rigs are stationed or are they simply anywhere oil is under the surface?
Would this mean Mobile bay is twice as affected due to the estuarine conditions and the number of oil rigs just outside the bay?
Are each of these enzymes found in all the Halo- genera or only certain species/specific conditions?
The soil DNA kit is obviously a good fit for this experiment as they are testing soil from multiple countries, but what other kits would be suited for this lab? Would a different kit have a major impact on the results?
I am unfamiliar with SDS-PAGE GELS so I looked into it. They are used specifically for proteins because the electric currents will only move the proteins based on weight, excluding other factors like charge. The charge of the gel itself blocks the charge of the atoms in the proteins and lowers pH. I also found out that Coomassie blue was an intentional choice as it is formulated work best in this type of gel.
What causes a lag period as mentioned here? Seminole degraded the given conditions but reasoning cannot be determined is the test inconclusive?
Do the pca and pob genes overlapping include each gene within that category mentioned or are is it variable?
In the Seminole strain the paa cluster reflects the halophile characteristic, can it also be present in halotolerant bacteria?
Since the 4-HBA grown cells are more adaptable to different compounds would that make any microorganism with these markers better suited for the degradation of carbon in the areas mentioned in the introduction? Or does adaptability have to do with the influence of other genes?
This sounds very similar to the previous paper based on the research on carbon usage and lack of previous knowledge. I looked into the bacteria being used and it is completely aerobic and has both terrestrial and aquatic species.
I am from Kentucky and it has a major coal/oil mining industry, specifically in the eastern part of the state. Many of the sources of pollution mentioned here are examples of what is being mined across the state. In the region where the most mining is the water is so polluted, there is always a boil water advisory because the treatment plants can’t keep up. I’m curious to know if research like this could be beneficial in keeping contamination levels down.
So far the methods of this paper are much easier to read and understand than the previous paper. Scientific papers, in general, are difficult because of the language used but after doing a lot of these procedures the writing makes more sense. I think I like it more because I know more about the topic.
I was curious about the difference between the gels we have run in class and the ones mentioned here. After some research, I found the gel is similar but the placement in the chamber is different, and therefore the way the electric current flows through it is different. The gel has each end in the buffer but the middle is not covered. The electrodes are on the top and bottom with the buffer and as it runs the lines will move from top to bottom.
Looking at the table I noticed that the % identity of the different orfs are much smaller than what we see in class. Is this due to the type of software we have access to or is it a characteristic of the strain used in this paper?
I am a bit confused about table 2. I see that the last two columns are indicating if each substrate can be transformed by DFDO or CARDO. is the % yield referring to the average when it reacts with both or is it referring to something else?
A lot of the nomenclature in this paragraph is a bit complicated to follow so I did some googling to better see what some of the compounds are. It seems that most are associated with either some type of oils or additives in insecticides or pharmaceuticals. This made more sense seeing as the testing focused on pah degradation.
If FlnA1/2 can both produce metabolites of DFDO and CARDO then would one be considered “better” or the same?
I also looked into the Glimmer program. It’s used for prokaryotic DNA to find coding and noncoding regions and focuses on long ORFs. It uses a few Markov models to run its data which is different from the other systems.
I agree, I think that the knockout could be useful but a PCR seems to yield a better result with more confidence. I would be concerned about environmental factors or mistakes more in a knockout than in a PCR test.
Could then the bacteria being tested under lab conditions act differently when in natural seawater due to the additional particles?
It seems the B30 strain is very different from the P73 and it does not degrade aromatics as well. It also has less abundant processes and shares no homologs. It’s interesting to see how different species can be even when a part of the same genus.
Can we discuss this section and or figure 5? I am not sure what regions they are referring to within the image.
does each arrow in the figure represent one gene found or is it a representation of an orf/predicted gene?
I knew what HGT was but I was unsure as to how common it is. The paper mentioned how many genes were HTG and I was curious if this was normal or high. I found that in bacteria and other prokaryotes it’s very common for this to happen and will often make up a large portion of the genome.
Even though this is in the results can this be discussed in the intro as previous studies since it is relating data to the other strains research?
How rare is it to find the first of something in the gene-sequencing world? I know much of the technology is relatively new so is finding the P73_0346 gene a major landmark or is this common due to the rapidly growing resources?
Could the ability to degrade fluoranthene so well along with the other aromatic compounds be due to it evolving to the environment it was taken from? The Indian Ocean is one of the most heavily polluted areas of the world, especially compared to the B30 strain taken from the Artic.
I want to go into physical oceanography and do research on pollution and or climate change affects. This study focusing on marine based bacteria could very useful in expanding knowledge on how to fight the ongoing issue of mass pollution. If there are natural ways to reduce pollution instead of using man made things with byproducts that are just as bad we can finally find a solution.
I think there’s a lot that can be learned from deep sea sediment and from sediment farther below the surface, especially in regions like this where the water is so old. I think studying bacteria near hydrothermal vents would be interesting as well due to the unique conditions and food sources available
What type of environmental conditions impose selection pressures?
Why does the biological method take longer than the physicochemical treatment?
After reading the entire paper, yes the biological method takes longer, but the results are better. Quality over quantity.
Is the purpose of transporting the samples on ice to prevent contamination and anything from being altered?
I did some research on this because I was also curious as to why it happened and this process is called thermocycling.
Is the cloudy white substance fungi?
So bacteria that exhibits high levels of degradation will also have high cell surface hydrophobicity?
In the previous section I asked if there was a correlation between emulsification activity, cell surface hydrophobicity, and naphthalene degradation and now I see that there is.
I believe it was just worth noting the variation because in the results section it stated that only eighteen showed an adequate amount of growth and were used for more study.
Has this process been beneficial or started any other studies in different locations?
Is it true that the presence of two or more benzene rings allows them to disperse at a wide range?
I found it interesting that Cycloclasticus means “ring breaker” and this ability plays a key role in their degradation of PAHs.
The flanking region contains regulatory sequences and is not transcribed into RNA.
I also did not understand this so I attempted to look up an answer or at least try to gain a better understanding, but I had no luck. Could someone please explain this to me?
In the previous section, I made a comment about sulfide because I did not understand why it was mentioned but now I see that it can be used as an electron donor.
Short Term: vomiting, diarrhea
Long term: cancer, damage to organs
I read that places such as Ireland do routine checks on PAH levels by taking samples of products that are suspected to be contaminated and then analyze them.
What is the method of Marmur? I understand that it’s used for isolation, but how does it do so?
It contains DNA fragments of a genome from a single organism.
Sau3AI is used because it recognizes the sequence GATC, correct?
Isn’t this a good thing?
I believe the upper pathway is when the aromatic ring is destabilized and the lower pathway results in the production of metabolites that can be used by the bacteria to form biomass.
Inorganic arsenic compounds are more toxic than organic compounds because they are more rapidly absorbed.
One source stated that if primers are unfit for an environment it will result in the inability to obtain PCR products.
Why were the slurries stored in the dark?
What role does sulfide play in arsenic metabolism?
Anoxygenic Photosynthesis requires light.
wasn’t this done in order to determine the electron donor?
Has anyone done a follow up experiment?
A monophyletic trait is one that is shared among closely related organisms. And it makes sense that arsenate reduction is not a monophyletic trait. This trait could have possibly been a result of convergent evolution.
This sentence was a little confusing for me. Can someone clarify? I basically understood that isolating the PAH-degrading bacteria that are native to Antarctica can help for treating contaminated soil both in Antarctica and elsewhere in the world. Is that correct?
I’m pretty sure the different levels of PAHs that you’re thinking about are more like different sizes/different number of benzene rings. If you search a picture of phenanthrene, you’ll see it’s a structure of three benzene rings. People are just interested in these because they are a large percentage of what makes up diesel-fuel.
Why was the surfactant production an important thing to test here? Does the production relate to the ability to degrade phenanthrene somehow? Perhaps it affects the ability of the bacteria to swim to its target?
You’re right, it is weirdly like a gram stain. I don’t think it’s meant as a real gram stain because they only used the one dye and never used a decolorizing agent. I assume it just dyed all the bacteria in the culture and they measured the absorbance of the crystal violet to measure the density of the cells in the culture. Almost like a using simple stain.
When it mentions that D32AFA did not differ from the control, it mentions that it means it cannot adhere to the phenanthrene. What are the repercussions of this? Is it possible that this is why it has the lowest degradation of the three strains chosen?
Based on figure 4B, the concentrations of Cd in the Antarctic soil would render the microbes practically useless for degrading phenanthrene. I’m surprised that wasn’t mentioned outright in the results. I wonder if it’ll be in the discussion.
I calculated the lowest degradation % in Figure 4B to be 5mg/Kg if were assuming 1mL= 1g. Considering the concentrations in the snow was 15-85mg/Kg, it’s unlikely that the microbes can degrade at all in the Antarctic soil.
I think that the extreme environments require the microbes to be more innovative with their uptake of nutrients so researchers are more likely to find the unique metabolic properties in those areas.
In order to decontaminate the oil spillage areas through bioremediation, do they just plan on growing specific microbes in a lab and dropping a lot in the area to clean it up? Similar to probiotics?
I would have never guessed oil would make it to the depths of the seafloor! It’s crazy how much an oil spill can affect everything.
So in this case of SIP, would they be creating oil with an isotope that is different from everything else they give to the microbes and see if the isotope ends up in the cells?
[…mineralization assays using 14C-labeled…]
What’s the difference between performing mineralization assays using a heavier isotope and SIP? I thought that’s what SIP was.
I understand the need for an internal standard, but if you’re adding unlabeled E. coli DNA to a mix of enriched and unenriched sample DNA, wouldn’t the E. coli DNA get confused with the unlabeled sample DNA and possibly mess up the measurements? With measurements as small as 5 microliters, it wouldn’t be very accurate.
Or was this separation not measured and used specifically for visualization?
When I originally looked at Table 2, I was confused to see 4567-24 showing higher mineralization yields than sample 4571-2. I’m glad they touched on why that may have occurred. Would it be realistic to see a future study where samples were taken in the same location as 4571-2 and tested using the same method except under anaerobic conditions?
So, on the DGGE, the top bands are the ‘heavy’ DNA and the bottom are the ‘light’ DNA, and the fact that samples 6-10 show just ‘heavy’ where 12-17 show primarily ‘light’ DNA shows that the DNA was properly separated through isopycnic ultracentrifugation. Then they used samples 6-10 for further analysis because they had the ‘heavy’ DNA.
Is that correct?
I agree. They took the 4571-2 microbes from a hot, pressurized, sulfide-rich environment, then tested them under conditions better suited for the 4567-24 microbes. I’d like to see another experiment with conditions more similar to the 4571-2 environment.
Agreed. It seems so much harder to get energy out of PAH’s than other carbon sources. They must not have anything else around to take from if they lost their ability to get energy from other sources.
Are these chemoautotrophs using As(V) as their electron donors and energy source, then CO2 as their carbon source?
Does the soil in Japan naturally have higher levels of arsenic in it? Is that why even uncontaminated soils have microbes that are capable of this? Or is this normal, and they were just giving an example from Japan?
What was the purpose of the RFLP? They mention further down that it was for grouping, but I don’t understand how that helps with grouping.
I believe so. They figure out the chemical makeup of the water, then recreate it under lab settings to ensure it stays the same and stays sterile throughout the experiment. I’ve done it before with tapwater experiments.
Either that or the abundance of microbes would adjust so the microbes that make As(III) would be more abundant than the ones that use it in order to maintain the concentrations.
I’m curious why the temperature range was wider at night than during the day. I expected it to be lower because it’s colder at night. I guess maybe it’s just circumstantial that it also deals with high temperatures better? Or maybe I just don’t know enough about hot springs to make an educated guess.
It’s cool to see that they can use either hydrogen or sulfide as electron donors for their reactions. It’s even cooler that we now understand why hydrogen works so much better for microbes as an electron donor!
This paragraph sounds like there needs to be a lot of future research on differentiating the bacteria in these samples. They can tell you that some bacteria are similar to previously published ones, but they can’t tell you which ones or even confirm that it’s the bacteria of interest that are related.
What was the purpose for using multiple kinds of antibiotics?
Why wait for five days? Shouldn’t they obtain a next-day result?
PAHs are then transformed through different peripheral pathways into a few key intermediates (such as protocatechuate, salicylate, gentisate, and catechol)
Have we talked about these before? I don’t think I remember these in the previous papers, either.
I think I am confused as to why it seems that they are studying this bacterium and this specific PAH when they seem to have already pointed out how similar they are to their respective counterparts.
I agree with Nevil about making mutants. It seems to give quick and easy results.
Is this an expensive process?
Is this to improve regulation?
Maybe they were under a time constraint or like it’s already been mentioned, maybe they were limited in their research.
I’m curious about this also. Is this a unique characteristic?
If I remember correctly, cosmids are can hold more than plasmids can. Maybe this is why they used them.
Could you add a substrate and the needed binding sites to make the gene work, or would it be easier said than done?
Regulatory genes are often transcribed in the opposite direction. They are often on separate transcriptional units (makes sense given that they control the transcription of other genes) and having them transcribed in the opposite direction helps ensure that they are regulated independently of the genes they regulate.
They want to examine the roles of particular combinations o genes so they have to subclone them. Look at the gene map figure to see which pieces they’re putting together.
This is why we make you take organic chemistry!
Ashlyn asked “So, are they expecting to find multiple genes in this study involved in the degradation?”
Ashlyn asked: So, are they expecting to find multiple genes in this study involved in the degradation?
Aliyah comments:
” However, it was recently reported that the LysR-type transcriptional regulators (LTTRs) ThnR, PcpR and LinR, which activate the expression of target degradative genes, were isolated from Sphingomonas strains that degraded tetralin, pentachlorophenol and γ-hexachlorocyclohexane, respectively.”
I read the other section connecting to this and cannot understand the purpose of including this sentence. Can it be explained?
Aliyah asks:
“We also investigated the regulatory mechanism of a newly isolated gene cluster…”
Is this somehow tied to the transcripts mentioned earlier?
Ashlyn commented: This portion basically answers my first question, but I am interesting in seeing exactly how many genes are involved in the degradation process.
Sierra commented:
“However, the genes for the intrinsic electron-transport protein for ArhA1A2, the genes for the degradation pathway of their metabolic products, and their regulatory genes, have not been isolated.”
In the sentence after this, they talked about using transposon mutagenesis for isolation except for that of Arh1A2. Why is this?
Sierra commented:
“We also investigated the regulatory mechanism of a newly isolated gene cluster, which includes the arhA genes, as the first step for elucidating the entire acenaphthene degradation system in strain A4.”
I thought in this particular kind of bacteria the genes were scattered throughout the genome. Were the genes clustered together in the lab, or am I getting this confused with something else?
I’m not sure why acenaphthene is less studied than naphthalene, phenanthrene, and pyrene. It’s on EPA’s priority pollutant list. It may be that it’s more difficult to obtain isolates and that the “true” PAHs are easier to study and a better model for aromatic hydrocarbon biodegradation. If you look up the MSDS for acenaphthene and compare it to naphthalene, they’re pretty equivalent.
Most bacterial groups other than the sphingomonads co-locate their PAH degradation genes. The sphingomonads are unusual in that their PAh biodegradation genes are not all located together in the chromosome. The sphingomonads tend to have a lot of small groups of PAH genes but don’t usually have the entire pathway at the same location. It makes finding the genes for the entire pathway more challenging than for say, pseudomonads. One explanation for this is that the sphingomonads may undergo more genomic rearrangements than other genera of bacteria.
The sentence after next gives you the answer to your question. Draw out the sequence of experiments if you’re still stumped.
That’s not really important. Focus on why they did the experiment. What did they hope to achieve by doing transposon mutagenesis? Were they doing random or targeted mutagenesis? What genes were they hoping to disrupt and why?
They did two separate disruption experiments, one where they knocked out ORF 15 and a second where they knocked out arhR yielding two mutant strains A4DR (no arhR) and A4D15 (no ORF 15). THe direction of transcription is irrelevant for the knockout as they did the knockouts separately.
Why do you think they chose to knock out these particular two genes?
Here they are expressing the dioxygenase genes (arhA1&A2) with and without the A4 electron transfer proteins (ArhA3 and ArhA4) to determine which gene products are necessary for dioxygenase activity. Remember that in our first paper they used electron transfer proteins from another Sphingomonas strain and did not get any indigo formation when they expressed arhA1A2 alone.
By amplifying across 2 genes they get to see if the genes are transcribed as one polycistronic message (controlled by one promoter) or if they are transcribed individually (each from a separate promoter). This experiment allows them to assess regulation of the acenaphthene degradation genes.
See the first paragraph of this section – Gm is short for gentamicin, an antibiotic. The miniTn5 transposon they used for random mutageneisis contains a gentamicin resistance gene as a selectable marker. All mutated A4 colonies will acquire Gm-resistance when the miniTn integrates into the A4 genome.
Kristen is correct. They did mutagenesis and some mutants lost the ability to convert indole to indigo. This ability is a proxy for the first step in PAH degradation. The mutants that can no longer do this must have a mutation affecting the initial step in the pathway.
Mutants that no longer cleared acenaphthene from the plate could have a mutation anywhere in the pathway before the metabolites enter central metabolism.
You need to think through/remember what you learned about PAH metabolism in the previous papers.
They know which genes are missing in the mutant and which ones are still present. By adding back some of the missing genes they can assess their function more accurately than in E. coli as all the other acenaphthene degradation genes are present and should be transcribed and translated correctly by A4’s genetic machinery, something that isn’t guaranteed in E. coli.
Does everyone understand this part? If you do plasposon mutagenesis as part of your project you will tkae a similar approach to “rescue” the mutated region of genomic DNA..
Exactly – larger inserts mean the investigators can screen fewer colonies and still be confident of covering the entire A4 genome.
The %identity is quite high for proteins. They’ll do experiments to get further support for the functional assignation.
You could if you wanted to.
The interesting thing in this section is that they find a number of pseudogenes that are truncated, suggesting the A4 genome is pretty dynamic and that rearrangements sometimes lead to formation of these truncated genes that are missing regions that are necessary for activity.
They can’t show all the data and this information is not critical to the “story” they’re telling in this paper. You don’t have to show all the data. Sometimes editors make you eliminate some figures because they’re not essential.
What you should focus on here is understanding which clones can form indigo and which can’t and why. What genes are essential for indigo production from indigo?
Pay attention to figure 1 as you read this section.
It allows you to understand the regulation of the acenaphthene degradation genes. They can see where RNA polymerase biinds. They can examine that region for other regulatory binding motifs. They found an activator and it’s binding site but there could also be regulation by a repressor protein (think of the lac operon and how it is regulated by both an activator (CAP) and a repressor (LacI)).
It may help to recap basic PAH metabolism to understand how to iinterpret the mutant data. Think about the initial steps in PAH degradation (it may be helpful to review Figure 1 from paper 2). The first step is catalyzed by the dioxygenase enzyme (with help from ferredoxin and reductase) and converts the PAH to a diol by adding both molecule of molecular oxygen across a double bond in one of the rings (acenaphthene is converted to acenaphthene cis-1,2-dihydrodiol and to 1,8 acenaphthenol by a subsequent step). This is the dioxygenase can also convert indole to indigo.
Based on this knowledge a mutant that can no longer convert indole to indigo must be mutated in a limited number of genes. Make a list of all the possible genes that could lead to an indigo-minus phenotype.
After the initial dioxygenase steps there are a series of reactions (~5-7 steps) before the PAH is converted to a central metabolic intermediate that can be oxidized to yield ATP/NADH/NADPH to fuel growth. Mutations in any of these later steps would be able to produce indigo but would not be able to grow with PAH as the sole source of carbon and energy.
Inducers are effector molecules that bind a regulatory protein to alter it’s activity. to alter arhR expression we’d need to look at the regulatory mechanism that contrls transcription of arhR.
One possible explanation for the constitutive expression of arhR is that there has been some mutation in the arhR regulatory regions (promoter/operator) that have lead to continual expression of arhR.
Or, it could be that arhR is continually expressed by design. Remember that the lac operon is continiually expressed at a low level (necessary so that when lactose is present LacZ can convert it to the induce allolactose) but it’s transcription increases 1000-fold when the inducer allolactose is present.
I would say that it’s still worthy of study – after all it works! In class we learn about models like the lac operon that are often quite simple (and efficient) but don’t always represent what happens in non-model systems. What we see in A4 is that the genome is dynamic and constantly being remodeled by transposition etc. Sometimes this is advantageous, for example when A4 acquires useful genes such as acenaphthene degradation genes. Other times it can be deleteroius, such as when the transposition event yields pseudogens that lack essential prts of the gene (such as in ORF11).
I think it’s likely that other regulatory elements may come into play. This is a catabolic operon and may be subject to both positive control (by ArhR) and negative control (by some as yet unknown regulatory protein, maybe a repressor-type).
It’s mostly a lack of studies, although in my opinion the authors are overstating this “lack.” There are quite a few studies on biodegradation of PAHs by marine bacteria and the pathwyas to date are quite similar no matter where the bacteria were isolated (terrestrial vs. marine).
This is a reasonable assumption. If you look at the response of the Gulf of Mexico bacteria to the Deepwater horizon spill it was clear that the bacteria were already primed to degrade the oil from both years of small scale spills and the existence of many natural oil seeps in the Gulf.
I think the goal is more to understand the mechanisms/pathways for PAh biodegradation rather than to develop commercial super-degraders, although there are some commercial strains on the market for biodegradation of chlorinated hydrocarbons. The chlorinated compounds are much more persistent than PAHs so it makes more sense to add specialized degrading bacteria to those spill sites. PAHs are degraded on much shorter time scales naturally and the most you can do to enhance degraration is to add nutrients (N-P-K, like for the plants in your garden) and maybe a co-metabolite (easier to degrade carbon source to get the bacteria started).
Grammar note: bacteria =plural, bacterium = singular! The genus Celeribacter is a member of the Rhodobacteraceae – you can just google this
I believe that the LB126 paper may have alluded to this. These are all mono-aromatic rings that are central intermediates in the degradation of a variety of PAHs. These intermediates are catabolized by a “lower” pathway that converts them into TCA cycle intermediates. The “upper” pathways are more specific to the PAH (e.g. phenanthrene vs naphthalene vs fuloranthene) whereas the “lower” pathways are more universal.
See my reply to Hampton’s comment above.
You’ll have to look up the answer in the references. I’ve posted the reference list separately.
They have the full genome. They told us that in the previous paragraph and in the last paragraph of the introduction.
Probably because they’ve used it before. you should look up how it works.
Focus on what’s in the main paper and on genes that may be related to PAH biodegradation. Otherwise there is too much information to process.
The cost is down to about $250 for a bacterial genome so not too pricey for a well funded lab. The advantage of doing the full genome sequence is that you’re not biased by previous research. You can (in theory) find novel genes that haven’t previously been described (especially if you use techniques like microarrays or proteomics).
GIs are regions of gDNA that look like they were acquired from an unrelated organism. They are often identified by looking at %GC as this % is usually the same value throughout a bacterial genome. If you see a region of gDNA with a markedly different %GC it was probably acquired by HGT.
In the next section they’ll also present all the potential PAH degradation genes they identified though sequence homology. This is essentially a list of potential targets for directed mutagenesis.
It does. Based on the metabolites they can tell what kind of enzymes C. indicus possesses and what knids of genes they should be looking for.
Given that you have the entire genome sequence would you use RT-PCR or another technique to look at gene expression?
We’ve been reading about “oddball” sphingomonads which undergo a lot of genomic rearrangements. It’s more typical to have all of the PAH degradation genes next to one another.
It’s hard to say with out looking at the entire genome sequence. It may be that C. indicus has pathways for synthesis if all the amino acids while B30 doesn’t. It may be that C. indicus evolved in an environemnt where PAHs were abundant and that over time it has lost amino acid transport & metabolism genes (relative to B30) because the supply of PAH was so high that it didn’t need to metabolize amino acids.
Or, the differences in gene content could simply be random and have no effect on the relative fitness of the two strains.
They don’t tell us where the genes are (chromosome vs plasmid) yet. We’ll have to read on. The big thing to notice here is that there are a lot of genomic islands in the P73 genome. It seems to be a strain that is good at acquiring and integrating foreign DNA into it’s genome. It may have some of the DNA uptake proteins we discussed in BLY 314.
This paragraph tells us that P73 is metabolically versatile. It can take up a lot of different sugars and feed them into multiple catabolic pathways.
Yes, they’re talking about HGT here and HGT from different sources. There have likely been multiple events where P73 took up and incorporated foreign DNA – both plasmids and the genomic islands discussed earlier.
It’s possible that they were lst somehow, especially given that so many other chemotaxis and flagellar genes are present. They don’t tell us here if the che & mcp genes are in the core genome but they probably are.
They have the entire genome so they’re likely to find more genes just because they’re looking at way more DNA sequence. They’re also lucky in that the genes are located next to one another unlike in many of the Sphingomonas species we read about in our earlier papers.
If it still had a functional flagellum it could move towards teh source of PAHs. Remember how we looked at regulation of chemotaxis and flagellar motion in BLY 314?
Similar gene arrangement (synteny) along with a high degree of sequence identity would suggest a common origin for the gene cluster.
These matabolites are produced during metabolism of a variety of PAHs. For example, some bacteria metabolize naphthalene with phthalate as an intermediate, others proceed through a salicylate intermediate, while still others go through gentisate. It’s a bit strange that P73 shows genetic evidence for many different PAH degradation intermediates but given the amount of HGT it seems to have undergone maybe we shouldn’t be surprised. It may have acquired PAH degradation genes from a variety of sources, each of which used a different “lower” degradation pathway.
Think about the arrangement of the lac operon. What genes does it contain other than beta-galactosidase? Is one of the genes a lactose transporter gene? Why would it make sense to have the degradation and transport genes in one operon? under the control of one promoter?
kan = km = kanamycin resistance gene. They used it because P73 was susceptible to kanamycin.
Yes. they’re based on the bioinformatics methods described in the methods and SI.
They tell us in paragraph 35 that it cannot degrade naphthalene, phenanthrene, pyrene or fluoranthene.
I’m not sure that I’d say that the genes are “new.” After all, they wrere identified by their similarity to previously described genes.
I think I’d phrase it the other way around – that the ability to take up a wide variety of compounds helps P73 degrade those compounds – I’d consider transport/uptake the first step in degradation
They could, although they may not have seen such clear results. Given that they identified a number of candidates that would facilitate PAH uptake it’s likely that there is some degree of redundancy and that you wouldn’t see a clear loss of function the way you do when you knock out the gene that encodes the first step in the degradation pathway.
There are other techniques that they could assess whether genes are linked to growth on fluoranthene – can you suggest any?
They probablyused either %GC or codon usage to identify the PAH degradation region as “foreign” or acquired.
They found putative PAH degradation genes in four regions of the genome and labeled them regions A, B, C, and D to make discussing their findings easier. It’s just an operational definition rather than a concept you should remember from BLY 302/314!
This is true. In many ways I think genome papers are boring. They tend to be very descriptive, reporting on a laundry list of genes and what they might do, with very little experimentation. This paper is actually pretty good in that they did some directed mutagenesis experiment to test some of their bioinformatics predictions.
It’s really the reverse, where the information from papers like our previous papers allow the computational predictions. We need papers where gene function has been verified experimentally before we can use sequence homology to predict function as they did in this paper.
The significance is that in contrast to other strains which seem to add the oxygens at multiple positions, P73 only utilizes one option (addition of oxygens at carbons 7 and 8). This should make elucidation of the subsequent steps in the pathway easier as there aren’t multiple degradation pathways being used at once.
Yes, because they have the entire genome they can identify all the potential PAH-related genes computationally and then map functions on them through trying to match metabolites to particular enzymes they detected computationally or by knocking out genes and seeing how the phenotype of the mutant differs from the wild type.
Contrast this to the approach you (and the first three papers) took where you had to spend a good deal of time simply finding a gene to test.
What genes were found on the plasmids? Were any related to PAH degradation?
In some ways this paper is a preliminary study. They sequenced the genome and discovered a large number of potential PAH degradation genes. They did some metabolite analysis to map functions to some of the genes and did mutagenesis to verify function of another. However, a lot more work could be done (verify function of the other genes).
The study of all the other PAH genes they identified represents probably 3-4 more papers. We can’t expect them to do it all in one paper. For one thing, many journals have page limits for their articles. For another, this article already presents us with a lot of information (especially if you read the 129 page supplemental information!)
This is something you should look up. I’ll post the references so that you can check the references cited to support this claim. Also, try to focus questions on the microbiology aspect of the paper. While the harmful effects of naphthalene etc. on mitichondrial respiration is important we want to focus on the isolation and characterization of bacteria capable of degrading PAHs.
We’ll find out as we move through this paper how long the isolates in this study take to degrade naphthalene. It depends based on the composition of the contaminating material.
Yes, the larger PAHs are more stable and more resistant to biodegradation.
Probably, but not necessarily in a negative manner.
Who/what do you mean by “those”? Any organism with mitochondria could potentially be harmed if the dose was large enough. It’s usually the larger PAHs that are harmful, not naphthalene. Naphthalene is the active ingredient in mothballs which you can buy and WalMart and use to protect your clothes. It’s harmful if you ingest it but you’d have to inhale a really large dose to suffer harm.
The authors are referring to damage to eukaryotes in PAH-contaminated environments. You’re correct about the difference in location of respiration in bacteria and eukaryotes.
It depends on the bacterium and the particular PAH. If you calculate the energy yield from the oxidation of a molecule of naphthalene all the way to CO2 and compare it to the energy yield for the oxidation of a molecule of glucose all the way to CO2 you’ll see that naphthalene is a pretty good growth substrate.
Combustion is one possibility. You send the contaminated material to an incinerator. You can also do chemical oxidations such as the Fenton oxidation where iron ions are used as catalysts.
Bioremediation happened naturally in the Gulf of Mexico after the Deep Water Horizon blowout. Naturally occurring oil-degrading bacteria bloomed in response to the released oil and degraded a large fraction of the oil and gas that was released into the Gulf. Most of the time you don’t have to add bacteria, they’re already there. you might have to add some fertilizer because the oil has a lot of carbon but lacks trace nutrients (N, P, K etc).
Why do you transport or store perishable food items on ice? To slow down microbial activity. Why would they want to slow down the activity of their samples?
They’re making stock cultures that they can use in the future. Storing a culture in glycerol between -20 and -80C keeps the cells viable indefinitely. They can sit in the freezer for years. It means you don’t have to worry about transferring the cultures onto fresh media weekly to maintain viable stocks.
I don’t understand your question.
For the first part, what drives phenotype?
For the second part, the authors just told you they chose phenotypically different colonies, colonies that have different appearences. They don’t want to isolate the same species 20 times, they want different species.
We’ll talk about this in a few weeks and do PCR in lab later this semester. Basically PCR is a way to make many copies of a piece of DNA from a target. It uses 2 oligonucleotide primers and a thermostable DNA polymerase. Because you need single stranded DNA to carry out DNA synthesis you first melt (denature) the double stranded DNA at 94-100C. Next you lower the temperature so that the primers can bind (anneal) to the priming sites. Then you increase the temperature to allow the DNA polymerase to make copies of the DNA. You repeat this process 25-40 times to end up with millions of copies of your target. See these videos for more details: https://www.youtube.com/watch?v=nHi-3jP6Mvc or the video in the study area in Mastering Microbiology
The answer to your question is in a previous comment!
The other main feature of ONR7a is that it contains all the major ions found in seawater. It’s synthetic seawater. It’s also a defined medium, not a complex medium. It’s also selective because limiting the carbon and energy source to naphthalene means that only naphthalene degraders should grow on it.
The MSM (minimal salts medium) we used in lab and supplemented with naphthalene crystals is similar in that it serves as a proxy for soil/freshwater systems.
Yes they’re using an enrichment technique. By limiting the carbon and energy source to naphthalene they’re selecting against all the bacteria that can’t use naphthalene. See my reply to Bently’s comment on the previous paragraph.
These tests just tell us more about the bacteria and let us know if we isolated 54 different strains or if some of the 54 are the same strain/species. Before DNA sequencing of the 16S rRNA gene became the standard for identifying bacteria you would do a variety of staining and biochemical and physiological tests and compare them to results in Bergey’s manual to identify an isolate to genus and species.
You need to amplify or isolate a large amount of the 16S gene away from the genomic DNA before you can sequence it.
Think about the surface properties of the bacterial cell and how that might affect the interaction of the bacterial cell with naphthalene. Is naphthalene hydrophilic or hydrophobic?
Some bacteria may be adapted to growing on high concentrations of naphthalene while others are better at growing on low concentrations. This could have applications if you wanted to use some of these bacteria to clean up spills depending on the amount or concentration of oil or naphthalene spilled.
You don’t want any water in the sample you inject into the GC-MS. It would harm the instrument.
This is something you can look up.
Biology is variation! Think about our class. We’re all members of the same species but have vastly different phenotypes. Why should the bacteria be any different?
Perhaps. They could also be autotrophs or they may have been growing on waste products released by other bacteria on the original isolation plates and not on the naphthalene or they could be autotrophs. There are a lot of possibilities.
According to the 16S sequencing they’re different genera and they probably have different surface properties that account for the different BATH scores.
The red arrows are there to highlight the sequences that came from the bacteria isolated in this study.
I’m not sure what your question is. Do you mean the ancestor of all the strains isolated? Not all the species represented on the tree are naphthalene degraders.
Phenotype/function doesn’t really follow phylogeny in the bacteria, especially not for functions that can be carried on mobile genetic elements such as plasmids and transposons. A lot of genetic variation in the bacteria is transferred laterally rather than vertically. This means that the species a bacterial isolate belongs to doesn’t necessarity tell you how that isolate will behave.
That would be a flask with naphthalene and media but no bacteria. So any decrease in naphthalene concentration would be due to things like photooxidation, sublimination etc.
This is true and interesting. The naphthalene degradation enzymes, transporters etc. in each isolate clearly differ to yield such different degradation rates. The surface properties measured by the E26 and BATH tests are also a factor in the different degradation results.
Is naphthalene hydrophilic or hydrophobic? Would a more hydrophilic or hydrophobic cell surface “help” a bacterium attach to a naphthalene containing oil droplet and degrade the naphthalene?
It may be that above 400ppm the naphthalene begins to exert toxic effects on the cells. Up to 400ppm increasing naphthalene concentration promotes growth (more C,E, e- to fuel cell growth) but above 400ppm the harmful effects outweigh any benefit from the increased C/E/e- supplied by teh increased naphthalene.
They probably based this choice on results from previous experiments by themselves or others.
Size is not an issue.
I’m not sure but they can go up to at least 4 conjugated aromatic ring (pyrene and chrysene) and 5 rings (benz [a] pyrene).
Think back to organic chemistry. Why are aromatic rings so stable and why would compounds with more rings be more stable?
It’s a bit more complicated than the number of rings as PAHs with the same number of rings can have different stability depending on the arrangement of the rings for example phenanthrene is more stable than anthracene even through they are both 3-ring PAHs.
Yes, they started with 54 but they used the initial morphological,and biochemical tests to narrow the ones they studied in detail down to 18.
In your summary you should explain this process – how/why did they end up focusing on just 18 of the 54 isolates?
Yes and many research groups explored the diversity of oil degrading bacteria that grew up in response to the DWH spill.
Keep in mind that the teichoic acids are negatively charged. They probably wouldn’t enhance interaction with a hydrophobic substrate. Their function is to provide some rigidity to the cell wall and to bind cations such as Mg+.
The numbers are pretty constant for similar environments but the species composition will vary according to environmental conditions (nutrient rich vs nutrient poor, light availability, organic matter content etc.).
Destin is correct. They want to reduce their workload so that they can focus on the best degraders.
The Roling study didn’t isolate individual species, they examined the bacterial community that emerged in response to oil and identified them by sequencing 16S rRNA sequences obtained from the total community DNA, not from purified isolates. They didn’t directly link the genera identified in the community to oil/naphthalene degradation. This current study makes the link explicit by working with pure cultures.
You could use media that favor the growth of gram positives. You could also pasteurize the water sample. This would kill many gram negative bacteria and allow the gram positives to be more easily isolated, especially any endospore formers in your sample.
I think that one take home from this paper and the work we’re doing in the lab is that these naphthalene degrading bacteria are ubiquitous. If we use growth conditions that favor them (ONR7a or MSM + naphthalene) we can often isolate them. Thus, many environments are poised to respond to things like oil spills. We may not need to add any special bacteria.
It’s possible but it could also simply be a sampling artifact. If you were to search the literature you’d see that a wide variety of both Gram negative and Gram positive bacteria capable of PAH degradation have been isolated.
Remember the basic principles of evolutionary biology. The environment doesn’t cause anything. Mutations occur all the time in species. Some confer fitness advantages others are deleterious. In oil contaminated environments bacteria that had mutated to be able to utilize PAHs would be more competitive than those lacking that ability. The environment selects the fittest organisms but it doesn’t cause them. Mutations conferring the ability to degrade oil also occur in pristine environments but they probably wouldn’t persist in those environments because they wouldn’t confer a fitness advantage in that sitiuation.
Isolate or strain, not strand!
The bacteria can degrade a lot of oil but some components such as asphaltenes and resins are resistant to biodegradation so there is always a need for physical oil removal as we saw in the Gulf of Mexico after the DWH blowout.
Also, in some environments you need to add fertilizer (mostly N and P) for maximal degradation. This is because the oil is carbon (and energy and electron) rich but nutrient (N/P?K etc) poor.
It could cause all the effects listed in the first sentence of your comment. In fact, if you cooked your seafood on your backyard grill you might add to the PAH load of your food!
I don’t think you’d want to do this!
The goal of the paper is to answer this question!
We don’t know if they are.
We don’t usually add bacteria to an area. At most we add nutrients (N/P/K/Fe) to stimulate the growth of degraders that are already there.
This paper dates from the mid 2000’s when whole genome sequencing was still expensive and time consuming. Instead of sequencing a genome to find a gene you would usually fragment and organisms genome into smaller fragments and put it in a cloning vector (they use a phage system in this study) and transform the vectors into a host such as E. coli. The collection of E. coli clones was called a genomic library because it contained all the genes from the organism you were studying. It was easier to study the genes in E. coli than in the original organism.
Then all you had to do was find the E. coli clone that contained the genes you were interested in. How did they do that in this study?
It doesn’t matter. Focus on the question, not the technical details of a method.
If you want to understand the details of the methods take experimental genomics next semester!
You should be able to infer the answer from the text. They tell you what they used it for. Otherwise use google.
This isn’t really critical for our understanding of the paper.
Here they’re just describing the various techniques they used to manipulate and detect the DNA & protein they worked with in the lab as well as how they analysed the DNA sequences they obtained.
The dioxygenase enzyme needs a supply of electrons for activity. The ferredoxin and ferredoxin reductase ferry electrons from NADH or NADPH to the dioxygenase. There are many different ferredoxins that interact with a variety of enzymes in teh cell.
A subclone is a smaller part of the original clone, just like a sub contract is a small part of a larger contract.
They’re not trying to biotransform the cells, they’re doing an experiment to see which aromatic compounds (listed in table 2) the cells containing pPhnA can biotransform.
Look at table 1. It lists the genotypes of the cells and plasmids used. The pPhnA plasmid contains Ap-r which encodes for ampicillin resistance. You add ampicillin to the growth media to ensure the E. coli cells maintain the plasmid and express the genes it contains (including the cloned dioxygenase gene).
What they know here is that they found two clones that share a common Sau3AI fragment. One probably has more DNA to the left while the other has more DNA to the right but they share a middle portion.
They also know that both of the clones contain the genes necessary for the first step in PAH degradation (and maybe more steps, we’ll find out below).
To complicate things acteria sometimes use GTG instead of ATG. They use the DNA sequence rather than the RNA because they sequenced the DNA!
Because it is! The genes on a genome don’t all face in the same direction, although genes in an operon usually do, that way the proteins they encode can be translated off one polycistronic message.
The fact that orf7 faces in a different direction suggests that it may not function with it’s neighbours. It may be subject to different regulation. It may not eve be involved in PAH catabolism! The fact that it doesn’t have a “phn” name suggests it’s not involved.
Think about everything we learned about mobile genetic elements. These genes are often acquired through horizontal gene transfer on plasmids or transposons. Rearrangements can occur, especially during transposition. How would you look for transposon “footprints” in this region? You have the DNA sequence.
The identities are not bad for proteins! And the identities are probably much higher if we zoom into the catalytic and substrate binding domains. They wold probably find better matches today because the database is so much larger.
This paper was published in the early 2000’s. They’d probably find a better match if they redid the search today. You could perform it yourself and see! They gave us the accession numbers so we can pull up their DNA sequences from Genbank.
You need both subunits for activity. It may be that the small subunit is located elsewhere on he chromosome or that the PhnA1b is non functional. There could have been a gene duplication event that duplicated the large but not the small subunit gene. Given the conservation in the Rieske and substrate binding sites the first explanation is more likely.
Yes but it looks like it’s paired with a different large subunit, PhnA1. PhnA1b needs a PhnA2b small subunit. The gene naming takes a little getting used to and they’re a bit sloppy. PhnA1 should would more correctly be named PhnA1a.
The big thing to notice on the tree is that PhnA1 clusters with dioxygenases previously reported to initiate attach on PAHs while PhnA1b is very distantly related and clusters with dioxygenases that act much further down the degradation pathway.
Proteins with as little as 30% identity can catalyse the same reaction. The key is he identity at the catalytic and substrate binding sites. That’s why they zoom in on the Rieske and mononuclear iron sites in this paragraph.
The beta subunits are often not as well conserved as the alpha subunits so this result is not a surprise.
I would say requires, not allows. It’s using the oxygen to break a C=C double bond in the PAHs. It’s not using it as a terminal electron acceptor in respiration in this case. These PAH degraders need oxygen for both processes. The dioxygenase pathway can only be used if oxygen is available.
I’m not exactly sure what you’re asking here. 4 genes are necessary for activity: A1 & A2 for he dioxygenase and A3 and A4 to transfer electrons to the dioxygenase so that it can add two oxygen molecules across the C=C double bond and break the aromaticity.
It just has to do with the structure/activity of the dioxygenase enzyme. Some things fit in the substrate binding site some don’t.
THe substrates were fused, not the PhnC! Look up the structures of the substrates.
What did you learn about meta vs paras ubstitued rings in Organic chemistry? Electron donating vs electron withdrawing groups? Maybe someone else can help us out. You’ve covered this material much more recently than me!
Would it be better to make a specific ferredoxin and reductase pair for every dioxygenase the cell encodes or to have one pair that can partner with many different dioxygenases? Which approach would better conserve resources?
If the electron transfer proteins can partner with many different dioxygenases it would make sense for them to be regulated differently. THey’d meed to be controlled by multiple inducers, not just the inducer(s) controlliing transcription of phnA1phnA2.
Why do photosynthesisers release oxygen? Where does it come from?
Basically whether it has an enzyme that can oxidise arsentite (releasing electrons that the microbe can use for ???) or reduce arsenate (for example as a respiratory terminal electron acceptor).
You can look this up in our textbook.
Think about why the microbes are using arsenate in particular. Would they still use it under oxic conditions or would they have more energetically favorable alternatives available to them?
What is the electron donor in chemoheterotrophy? What are the possible electron acceptors (other than oxygen)? You need to think about which As species is oxidized and can act as an electron acceptor and which is reduced and can act as an electron donor.
There are plenty of planktonic arsenic utilizing bacteria. Here the biofilm lifestyle may allow aerobic and anaerobic arsenic utilizers to grow in close association and cycle AsIII and AsV between them.
Yes, but we’ll see anoxygenic phototrophs in this study so it will be photosynthesis with a twist. One of the differences is that the phototrophs will use something other than water as an electron donor during photosynthetic electron transport.
Yes. Look up the reduction potential values yo understand why.
This is something you could look up in a scientific database such as sciencedirect, PubMed, or Scopus and report back to us. Jest search for arsenic resistant bacteria.
Many arsenic resistant bacteria exist but not all can use AsIII and AsV the way the bacteria in this study do.
It’s just a physical gradient – grater distance from the source of the hot water with the resultinig temperature selecting different bacteria and/or archaea.
You can look this up in our text. You may have to look up the two prefixes separately.
Exactly. They want to preserve the communities in the state they were in when they samples so they lower the temperature to slow biological activity down and put them in the dark to halt photosynthesis.
They don’t want to kill the samples as they want to use some in future incubation experiments so they store at 5C rather than freeze them.
They probably can’t. More importantly N2 isn’t usually used as a terminal electron acceptor in ETCs (why not?). By saturating the bottles with N2 they eliminate aerobic respiration and keep the samples in a state close to their natural habitat before using in their experiments. It should keep the community very similar to it’s original structure and activity.
Shaking the aerobic tubes/bottles increases aeration of the cultures and ensures that oxygen doesn’t become limiting at the bottom of the tubes/bottles.
Which form of arsenic? You need to be specific about whether you are talking about arsenite or arsenate.
Here they are referring to libraries made by ligating an PCR product into a vector. You could also make a genomic library where you ligated fragments of genomic DNA into a vector (what awas done in our previous discussion paper).
Probably the second option – the gene is a novel one that the current primers do not match.
What temperatures did they measure when they collected their samples in Mono Lake?
It allows them to add a very small amount of radio isotope and track where it goes. It means they can leave the natural concentrations of AsII and V virtually unchanged.
There’s no antigen here, antigens react with antibodies, but you’re on the right track. The use of isotopes makes it easier to measure what is happening to the arsenic or the carbon radioisotopes that they add to the samples. A small amount of isotope gives a large easily measured signal so they can add a little isotope and track what happens to the C or As.
The spec method is easier and faster.
Right, it’s a highly reduced molecule so it can serve as an electron and energy source.
The dominant clone was highly related to a bug they previously isolated from this same location. They also tell us that this bug, Ectothiorhodospira, strain PHS-1, can carry out some of the same processes they were exploring in the experiments in this paper. This is an example of a case where molecular biology approaches and culture based approaches give the same answer about which bacteria are dominating a system. This doesn’t always happen.
Yes. They include a killed control in order to show that any changes in arsenic speciation were caused by living cells, not just spontaneous chemical reactions.
What biological process requires light? What biological processes can occur in both light and dark?
These are “batch” cultures. Over time waste products accumulate in the bottles, slowing down the cells.
Pose a hypothesis yourself! Do all species have the same growth optima? How many different species are in the incubations?
Look up the reduction potential of the various electron donors they tested to see which would generate the most energy for the cell.
They don’t tell us. You could do a lit search of the Archaea they found to find out.
They set them up that way for ease of analysis although not all the experiments started with only one member of the AsV/AsIII redox couple in the incubation. Once the incubations are underway we have a mixture of AsIII and AsV. Look at the 50h time point in figure 1 for example.
They ran it long enough to see all the AsIII oxidized to AsV and then for long enough to see all the AsV reduced back to AsIII. The biology of the biofilms dictated the length of the experiments.
I’m not sure what you mean by de-energize.
Yes, but what you need to do is identify what kind of metabolic processes could account for these observations. During what kind of metabolism(s) would a cell oxidize AsIII (arsenite)? reduce arsenate (AsV)?
Arsenate is reduced in the dark forming arsenite. Spelling matters!
What processes would drive AsIII oxidation under oxic conditions? If the rate is the same with and without oxygen do you think oxygen is involved in the process?
Apparently.
Hard to say. It could simply be that the bugs using the arsenite are increasing in numbers as the incubation progresses and therefore the AsIII is oxidized more rapidly.
Why? I’m not disagreeing but I’d like you to support your assertion.
If sulfides are converted to various thioarsenic intermediates will the sulfides be available to the bugs to serve as electron donors?
No, I think we’re just seeing a little bit of abiotic AsV reduction or just analytical error. The error bars on the measurements are pretty large. The AsV time zero and final time point for the no electron donor trace may not be significantly different from one another.
Think about “who” would use the acetate or bicarbonate and how they would use it. Remember, bicarbonate is essentially CO2 and will be processed by the Calvin cycle or some other carbon fixation pathway. Also remember that anoxygenic phototrophs can use a wide variety of electron donors and that microbes are versatile and may use a number of different metabolic strategies depending on environmental conditions.
What they mean by “not clear” is that they can’t tell which explanation is correct.
Can you think of a genomics or mutagenesis based experiment they could do to identify the arsentie oxidase gene(s)? This would be a great exam question.
Why? Should all the arrA genes show similar homology? They probably come from a variety of different Bacteria and Archaea in the biofilm community.
I agree with Miranda.
We’ll discuss some techniques in class on Tuesday that should give you some ideas on how to link particular functions to particular microbes in a communtiy.
Read on! Maybe the next paragraph will help.
The previous paragraph tells us that acetate was not used as an electron donor (and carbon and energy course). If the acetate was used as a C, e- and energy source in anaerobic conditions it could use AsV as the terminal electron acceptor (ie stimulate AsV reduction). But they did not see AsV resuction stimulated when they added acetate in the dark.
But, in the light they see acetate oxidation so in the light some process that removes electrons from acetate . . .
The authors added it to the bottles. It’s just water soluble CO2.
You ca look this up. Search “arsenic bacteria mono lake” or something similar.
You’ll see as the semester goes on that a lot of work has been done with naphthalene (2 rings) but not as much with the higher MW PAHs. Naphthalene is easier to work with (higher solubility) and much less toxic than the higher MW PAHs which is why we’re using it for our studies.
One thing to think about is whether there is any reason fluoranthene degradation would differ in marine vs. terrestrial bacteria. My feeling is the bacterial species may differ but that the biochemical degradation pathways should be very similar. We’ll find out.
It’s most likely that a ring hydroxylating dioxygenase initiates degradation just as it did for the PAH degraders we read about last semester.
That information is usually presented in the closing paragraph – read on!
True, oxygen availability will influence biodegradation but if we’re looking at aerobic marine vs aerobic terrestrial environments the pathways will probably be similar, although the microorganisms may be different.
They’re not clear on this here but presumably the isolate came from the aerobic layer of the sediments.
No. This section is just a resource if you want to look up any of the papers the authors cite in other sections.
Given that PAH degradation genes have been isolated from other microorganisms they use bioinformatic analyses to search for similar genes in the C. indicus genome once they sequence it.
This bioinformatic approach will only work if the C. indicus genes are similar to previously described genes. If C. indicus has a completely novel PAH degradation pathway they will need a different approach.
The previous paragraph describes annotating the C. indicus genome (inicluding, based on the last paragraph of the introduction section, PAH degradation genes). Given their research goals we can infer that they wanted to mutate (knock out) a PAH degradation gene using the cre-lox system.
It makes it easier to separate and identify the various intermediates in the fluoranthene degradation pathway. If we can identify all the intermediates we can figure out what enzymes are required for each step in the degradation pathway.
They probably used an enrichment culture technique where they put some sediment and buffer in a flask and added PAH as a sole carbon and energy source. That way, only PAH degraders would grow up. That is what the BLY 314 students did last semester to isolate the naphthalene degraders we’re working with this semester.
Exactly. If they knock out or inactivate a gene and the mutated bacterium can no longer utilize fluoranthene that would support the hypothesis that the knocked out gene is necessary for fluoranthene utilization.
The key s to read the figure title carefully and then look at the big picture for each ring. For example, we are interested in PAH degradation genes – probablt categorized as energy productin and metabolism, so we could focus on those colors.
We also know that non-essential (but useful) genes like PAH degradation genes are often acquired by horizontal gene transfer so we might also want to look at the %G+C and GC skew figures and see if there is evidence of HGT from te %G+C across the genome. Remember that organisms typically have a fairly constant %G+C and large deviations from that % are usually indicative of HGT events.
Right. Fluoranthene is too big to simply diffuse across the plasma membrane. It’s chemical properties would also make diffusion difficult.
I disagree. Looking at the circles that depict G+C content, is G+C content consistent across the chromosome? across the individual plasmids? Given that G+C content is usually fairly constant across a bacterial chromosome or plasmid and that in these figures G+C content is not constant what can we conclude about the evolution of the genome of C. indicus? Based on the figures do you think the genome we see “today” is the result of only vertical evolution or has horizontal gene transfer also played a role?
It may be that this organism doesn’t engage in gluconeogenesis. It may use other pathways or acquire the glucose it needs from its diet.
Not necessarily. They could if they were on the same plasmid or transposon.
Poorly characterized is where they put all the genes that could not be assigned a role based on homology to other genes in the databases.
Yes. This figure shows us that the RepA ad ParA proteins from the C. indicus chromosome and plasmids do not share a common evolutionary history. If they did they would all cluster together on the same branch of each tree. The trees are another line of evidence that the C. indicus genome has been remodeled by extensive horizontal gene transfer.
Given that the proteins from the various plasmids are related to a variety of different bacteria we can conclude that the plasmids all have different evolutionary histories and were probably acquired in separate events.
They’re closely related but one degrades PAHs and the others doesn’t. The comparison is analogous to comparing the genomes of two E. coli strains, one virulent, the other non virulent. It’s a way to identify all the genes that may be involved in PAH degradation, not just the ones that are similar in sequence to previously characterized genes.
You’d expect genes that were acquired from the same source to share a high degree of sequence similarity but also exhibit conserved gene order. The gene order can change over time if the organism you’re studying undergoes frequent genome rearrangements. All they’re doing here is showing us the degree of conservation of gene order between C. indicus and the two bacteria containing the PAH degradation genes with the highest degree of sequence identity.
This is ;\likely to be more influenced by the temperature that P73 inhabits.
It’s likely that this strain had but has since lost the fli genes. Given the amount of HGT we see in the P73 genome this isn’t surprising.
Some transposons have sequence preferences for the sites they will transpose into, others are less specific. I think they focus on B here because the thrust of the paper is identifying the PAH degradation genes. It may also be that they didn’t identify insertion hotspots in the other regions.
You’d want to look at RNA, not gDNA. You could also use Reverse transcription PCR to examine when the gene transcript is produced and link transcription of the RHD gene to the presence of different inducers/substrates.
You could do qRT-PCR to quantify expression.
You could also make an expression clone that contained the RHD genes and transform it into E. coli and see if E. coli gained the ability to metabolize fluoranthene.
Both! They’re using sequences reported in previous research for their analysis.
Yes. It separates the various metabolites and identifies them.
You could knock out each gene in the pathway separately and look for accumulation of metabolite from the previous step in the pathway.
See my answer to Bentley in the last paragraph.
It’s because the aromatic structure is really stable. Even though we often draw the benzene rings with discrete double bonds the pi electrons are shared between all 6 C in the ring and attacking the aromatic ring is therefore more difficult than attacking a C=C double bond in an alkene.
As you point out genes acquired through HGT could be detrimental if their acquisition disrupts normal expression of essential genes.
Yes!
Based on the annotations many of them do. Have a look at the degradation pathway figure. They’ve put the “code names/numbers” of the genes they identified above the various reactions. If you look closely, some of those steps are oxygenations.
They may be used in other pathways if they’re not used in fluoranthene degradation.
Radical reactions could work in an industrial setting but not in a cell!
Generally bioremediation for things like PAHs relies on naturally occurring microbes. We don’t usually add designer microbes to a PAH polluted environment. In contract, environments contaminated with chlorinated hydrocarbons do have “superbugs” added to them as the ability to attack chlorinated pollutants is not as common.
For PAH contamination we might add nutrients/fertilizers (N/P/K) to add the elements that are missing in the PAHs but necessary for biosynthesis.
One strategy is enough if oxygen is present. P73 can also utilize a variety of other carbon sources.
Until this paper was published all the fluorene degraders that had been reported in the literature were Gram positive which was a bit odd. There is no biochemical or physiological reason that this ability would be restricted to Gram positives. After all, Gram negative bacteria degrade many other PAHs.
The other routes appear to lead to dead end products that accumulate as they can’t be degraded further.
The biggest different is the ability to grow in 3.5% salt for marine bacteria. Other than that terrestrial and marine bacteria share the same biochemical pathways for the most part. Both environments contain a lot of proteobacteria.
Meta-cleavage refers to the position on the aromatic ring at which it is cleaved. In IUPAC nomenclature meta is at carbon 3, ortho at carbon-2 and para at carbon-4. See more on meta-cleavage here https://metacyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-5415
Note how the pathway ends with metabolites that can enter the TCA cycle, a common pathway found in most microbes.
I believe that GSL stands for GlycoSphingoLipid and LPS is liposaccharide (found in the outer layer of the outer membrane of Gram-negative bacteria) The latter you should remember from BLY 314!
I’ll let one of you look this up. A simple database search for Sphingomonas and PAH should answer the question.
Sometimes the initial dioxygenase has a much broader substrate range than the enzymes further down the pathway so you end up with situations where you have an RHD that can hydroxylate a variety of compounds but the products it makes cannot be processed by the enzymes catalyzing the subsequent degradation steps. For example they might not fit into the active site of the next enzyme.
You designed degenerate primers targeted to Acinetobacter RHDs last week.
Anyone have any ideas on why the authors used a combination of enzymes to digest the genomic DNA?
Exactly. They included restriction sites at either end of the PCR primers so that they could clone the RHD into an expression vector in the correct orientation. Why is that important?
Why do you think they isolated the proteins? Which cells do you think they grew up and extracted?
Did anyone look up the purpose of sonication?
Gene knockout is definitely a possibility as well. Usually a reviewer will want to see two separate experimental lines of evidence that a gene is involved in a particular process. Expression cloning and knockout are two possibilities. This papers utilizes a third approach (RT-PCR) to confirm the expression clone results.
Remember that the purpose of GC-MS is to separate and identify metabolites. The sample is heated up and compounds with different evaporation temperatures will separate at different points along the temperature range allowing for their detection.
Yes. It makes cloning the RHD fragment easier by reducing the number of clones they have to screen to find the RHD containing fragment. It also means they should only get white colonies as the vector should not be able to self ligate because the sticky ends from the two different enzymes don’t match.
You’re on the right track. The sonnication breaks the cells open releasing the cytoplasmic contents. They don’t know if the RHD proteins are transported out of the cell or not so they lyse the cells to be sure that they “see” everything on the gel.
Think about all the information they gave us. LB126 can grow on fluorene (as it’s sole C source) but it can degrade phenanthrene, anthracene, and fluoranthene as long as you also supply pyruvate as a C source. This means that LB126 needs a supplemental C source when grown on the other PAHs because it cannot make all the biomolecules it needs for growth from them.
Think back to when you studies metabolism (both catabolism and anabolism). What do cells use pyruvate for? What pathways is it part of?
Previous work had found these lower pathway genes in LB126 (reference 47) and they were most similar to genes from Sphingomonas KA1 and CB3. They’re pointing out that these new genes also have some similarity to Sphingomonas genes but the matches are not as good as to the Gram positive genes. Maybe the other sphingomonads also acquired genes from Gram positives???
Look at the yield column in the table. Substrates that yield a high % of product are “good” substrates. Does fluorene yield the highest % product? What product(s) are produced from fluorene? Do we see the expected product? Was there anything unexpected here?
Pyruvate can also serve as a precursor molecule for many important biomolecules. When they incubate LB126 with the non-fluorene PAHs they need to supply a supplemental C source that can be used for both energy generation and biosynthesis as they can’t metabolize the other PAHs all the way to all the intermediates they need for those purposes.
No. It just means that E. coli was making the proteins but not folding them correctly so they couldn’t attack fluorene. They had to change the growth conditions (grow at 42C instead of 37C) to get more of the protein make in the active form.
Maybe, , but maybe not. The structure of the enzyme is also important. For example you could have an RHD whose active site fits fluorene better than naphthalene and therefore degrades fluorene more efficiently.
It has to do with which carbons on the PAH are attacked by the RHD. Naphthalene, for example is attacked at the 1,2 positions by “lateral” dioxygenation. Dibenzofuran is attacked at the 4, 4a carbons in an “angular” dioxygenation. See this figure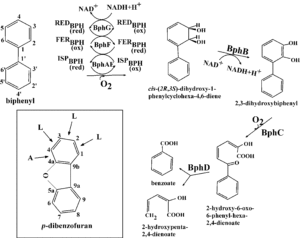
DFDO = dibenzofuran dioxygenase
CARDO = carbazole dioxygenase
Unlikely. We see very little HGT for essential traits like plasma membranes, DNA replication, cell division etc.
It’s hard to say as there is not as much work on acenaphthene and acenaphthylene biodegradationas on PAHs like naphthalene and phenanthrene. However, we’ve seen similar patterns in the previous two papers where one PAH can serve as a sole C ad energy source but other PAHs cannot (even when the RHD can attack other PAHs). It will be interesting to see if the RHD these investigators identify follows a similar pattern.
Probably! You can look this up on Google Scholar or PubMed.
They’re not exactly the same but they’re closely related. After all, in the last paper we read they used degenerate PCR primers that were able to amplify RHD genes from a broad array of bacteria.
From the studies they cite where they compared the PAH degradation genes they found in each individual study to all publicly available sequences.
Yes.
That’s right. Molly has more details above. They used it anytime they needed to cut a band out of an agarose gel and purify it so that they could clone it. For example you could digest genomic DNA, run the digest out on a gel, cut the size range you wanted out of a gel, and ligate it to a vector cut with the same enzyme.
Yes, using indole and looking for indigo formation allows us to detect the presence of an RHD.
Yes.
I imagine (but don’t know for sure because I’ve never used this system) that 1:1 gives the best results.
Yes, this is a good confirmation experiment and they confirm the knockout by Southern blot. PCR might be easier.
Yes, blunting does refer to removing the sticky ends. you’ll sometimes see it called “polishing.”
This particular experiment could be set-up better. We’ll see in the results and discussion why they need to find a spontaneous mutant.
They found two copies of the arhA1 gene (probably saw two different bands on a gel in a Southern blot with an arhA1 probe). So now they have to wonder if both copies are functional/active or if one is a pseudogene.
Exactly!
How so? Do you have an alternative strategy in mind?
Drawing a radial tree like this one is really just a matter of preference. Some people prefer this layout over the “boxy” trees we drew. MEGA lets you do either.
It’s a part of the protein necessary for activity.
They found two dioxygenase alpha subunits, arhA1 and a homologue when they did the Southern blot (described in the next paragraph).
They’re not referring to the gene used the homologous recombination part of the knockout experiment.
Ideally they’d clone and sequence the homologue so that they could figure out why it doesn’t confer dioxygenase activity. It could be a partial gene that has enough sequence to bind the Southern blot probe but is missing sequence necessary for dioxygenase activity.
It means that it’s related but not closely related which makes sense as acenaphthene and acenaphthylene are different enough from naphthalene and other PAHs that the acenaphthene dioxygenases could be expected to differ from the Nah-like RHDs.
Putting the RHD on one plasmid and the ferredoxin and reductase on a second plasmid means they could test a variety of RHDs in a host that contained the ferredoxin+reductase plasmid by just adding a second plasmid that contained the RHD genes. These genes are not always next to one another so making a clone that had RHD-alpha+RHD-beta+ferredoxin+reductase on one plasmids would be more difficult than using the two plasmid system.
There is at least one follow-up paper. We may read it as our last discussion paper. I haven’t decided yet.
They all share features like typical habitats, yellow pigmentation, and sphingolipids. Only some degrade xenobiotics.
You could use RT-PCR to detect expression of transcriptional regulators but more often when studying regulators you fuse the regulator promoter to a reporter gene and measure the activity of the reporter gene.
The gene clusters have promoters that are all switched on/off by the same effector molecule. Remember in BLY 314 when we studies regulation we compared the lac operon to the mal operon. In teh lac operon the catabolic genes are located together but in the mal operon they’re in different locations on teh chromosome but in both cases they were controlled in the same general manner (negative control of an inducible gene – repressor being inactivated by binding of an inducer molecule).
Today we’d probably sequence the entire genome. When this paper was published that would have been very expensive, hence they take a more limited approach.
They’re just pointing out a limit of their previous study and hinting that the found those genes this time. It could be that the ARhA1A2 protein will have even better activity with its true electron transfer partners.
Fixed.
Because it works. Sometimes electroporation or calcium chloride transformation don’t work so you try mating instead.
Yes! When you write your summary use the order of the tables and figures to help you put experiments in the correct/most logical sequence.
We’ll see that they use a variety of different plasmids and mutants and will need a variety of selectable markers to distinguish between them.
If you’re on campus you can access the journal. At home remote login to the library to see the tables. I’ve fixed the links so you can see the tables here now.
It works well for some strains that cannot be electroporated or calcium chloride transformed.
Measure cell density in a spectrophotometer and plot the data. When the curve plateaus the cells are in stationary phase.
They did a lot of subcloning using restriction enzymes. The TA cloning we did is more efficient (and cheaper to do than when this paper was published).
It “marks” the insertion – Gm resistant colonies are mutated, Gm sensitive colonies are not. It also allows them to clone out the mutated region using the Gm resistance as a selectable marker.
This allows them to identify the transcription start site which is useful in understanding regulation.
It helps here to remiond yourself what the first few steps in acenaphthene degradation are. 1,8NA is a downstream metabolite of acenaphthene. You could lose the ability to carry out step one ion the degradation pathway but still be able to do steps 2-10 if provided the substrate for steps 2-10 dpending on where the mutation occurs. Is it in the dioxygenase genes (disrupt step one but still have the ability to carry out steps 2-10) or is it in a regulator (lose ability to carry out any and all steps in the pathway because no mRNA is transcribed for any genes in the pathway).
Yes. We have to look up what each gene product does and think about how losing that gene would affect acenaphthene degradation.
They told us that the the substrate- and NADH-binding domains were deleted.
We’ll have to read on to find out! How would you answer this question in the lab?
Look at the genes that each expression clone contain. That should answer your question.
Yes.
It would make sense for arhR to be constitutively produced, right?
These are all good questions. generally we’re using bacteria that are “native” to the contaminated site or from a similar site so they don’t disturb the ecosystem. In terms of downsides bioremediation can be slow and can be limited by other nutrients such as N and P. This can be overcome by adding fertilizer to the oil spill area so that the oil provides the carbon for the bacteria to grow (and respire the oil to CO2)and the fertilizer adds the other required nutrients for biosynthesis of new cells.
This would be the best approach but is not always possible.
Typically they slow their growth rate substantially. Endospore formers can form endospores and wait for better growth conditions.
Think back to what you learned about aromatic and polyaromatic hydrocarbons in organic chemistry. Which do you think would be easier to break down, a 2-ring or a 4-ring PAH?
Some of the oil ends up as bacterial biomass and some ends up being respired to CO2. The ratio depends on the growth efficiency of the bacteria. Basically you see a transient bloom of bacteria that is then grazed by larger microbes so the oil carbon moves up through the food web with more and more being “lost” as CO2 at each trophic transfer.
Make sure your comment is tied to the correct paragraph.
It is a method for extracting and purifying DNA. If you want more information you should google it.
They’re not changing the concentration of bacteria in the environment in this paragraph. They’re just reporting where the obtained their samples and how they transported them.
Why do you store anything on ice or in a fridge or freezer? To slow down or prevent microbial growth. They want to preserve the sample “as-is” until they get back to the lab.
See above.
At that depth they’d probably only recover anaerobes or facultative anaerobes. Oxygen is depleted rather quickly in sediments
They only have so much time/money to collect the samples.
Focus on the experimental methods in your comments on this section. I want to make sure you understand the approach the authors took in their quest to achieve the goals outlined in the introduction.
Look up the recipes. One major difference is the amount of salt. Why?
I’m not sure I agree with you on this. What are their experimental objectives? In this section they’re isolating NP-degraders so they’re not concerned with replication or repeatability. They simply want to isolate as many different NP degraders as possible.
It has loads of salt (40g/L). Why?
What size are bacteria? If you filter seawater through a 0.2um filter and add the filter to some media what have you really done?
They’re measuring the density of the bacteria in the culture by measuring the OD (see the microbial growth chapter in our text). They’re not measuring DNA. For that you’d need to measure at much shorter wavelengths.
We’ll see in chapter 5 that there are other options but OD600 if perfectly appropriate.
You’d typically collect the same time points from every culture you’re testing.
They probably did streak plates where they put one isolate on each plate.
At the beginning they worked with 50+ isolates. This first method is faster and easier and accurate “enough” to allow them to distinguish between slow and fast NP degraders.
What do you think they should measure instead of the amount of naphthalene left (i.e. not metabolised)? Measuring the amount of NP left in the culture vessel is straight forward and gives you the amount used by the bacteria by simply subtracting it from the amount added at the beginning of the experiment.
There are lost of analytical methods to choose from. Our focus should be on why they made the measurement, not on how.
They have 50-something isolates. How will they figure out if they are all different or if some of them are different species? By doing morphological (staining), biochemical & physiological, and molecular (16S sequence) tests on them an d comparing the results.
We’ll send our isolates out to a lab that uses this kit to get teh 16S rRNA sequence of your isolates. In fact, we’ll do everything in 2.2.1 as well!
Your assumption is wrong for a simple PCR and sequencing experiment. We’ll talk about this later this semester
We’ll have to read on to find out!
What do you know about the bacterial cell surface (hydrophilic or hydrophobic)? PAHs (hydrophilic or hydrophobic)? What kind of cell surface would attract PAHs?
They gave you the recipe for ONR7.
The shaker allows for better air exchange so that the media remains oxygenated. What do NP-degraders need for the first step in aerobic NP degradation (I put this on the board in lab).
Google!
It was probably chosen based on the results of their earlier NP degradation experiments.
Make a guess. What could limit an organism’s ability to utilise a substrate?
This is like asking why your brother is taller than you are! If we had the time to really dive into this question we could look at things like differences in the gene sequences coding for NP uptake and degradation but lets just say they’re a lot of natural variation among bacteria.
They present some of these data later.
It keeps the cultures aerated. Remember thet first step in bacterial degradation of NP that I put on the board in lab? What was the other reactant in that first step?
You assign your isolate to a genus and species by comparing it to previously described species and based on the level of identity you assign your unknown bacterium to a genus and or species. For example, for bacteria, if the %identity of the 16S rRNA gene is greater than 97% we usually classify them as belonging to the same species.
If they only put their isolate sequences in the tree we’d be able to tell how closely related they were to each other, but we also want to include reference sequences so that we can compare them to the larger bacterial family.
Both the E24 and BATH tests measure how well the bacteria can attach to the hydrocarbons. The bacteria need to be able to “touch” the PAH to degrade it but most bacteria have hydrophilic surfaces and PAHs are hydrophobic so PAH degraders need to modify their cell surfaces.
All you need to know is that the GC-FID is more sensitive (but also more labor intensive and more expensive).
IS there a level at which bacterial activity seems to become inhibited.
As mentioned below it could be toxicity and probably is. If the bacterial transport or enzyme systems were saturated we’d expect to see a plateau, not a decline.
Look at the % NP degraded after 15 days. For some isolates it’s almost 90%. if they incubated them much longer there would be nothing for them to grow on.
No. It is just natural variation. Just look at our BLY 314 class, we’re all members of the same species but we show a lot of variation in our traits. Why should the bacteria be different. The isolates in this study are more diverse than our class, with a number of different genera present so variation in the rate at which they degrade NP and their BATH and E24 results are to be expected.
Not really. For the most part the two tests give similar results with the exception of isolate N4 which is a weird/unexpected result.
It basically would allow for the break up of large oil slicks into small droplets which would be more accessible to the bacteria.
This is a tricky question as you have t decide how to measure the concentration – what volume of seawater are you going to assess? The concentration at the sea surface or throughout the entire water column?
The more rings, the harder the PAHs are to degrade. Naphthalene (2) and phenanthrene (3) are degraded pretty rapidly, pyrene, chrysene, and benz[a]pyrene more slowly.
2-3 rings = degraded quickly (easily)
4 or more = degraded very slowly (harder)
If you do a search you’ll find a lot of papers describing naphthalene (2 rings) and phenanthrene (3) degraders but much fewer reports of pyrene(4) or chrysene (4) degraders and hardly any on 5-rings compounds like benz[a]pyrene.
You’ll also find a lot of reports on linear alkane degradation but few if any on branched or cyclic alkanes.
People have also looked at phenanthrene (3 rings), pyrene (4), chrysene (4) and found degraders. 5 rings or more tend to persist in the environment.
Yes and on land (like we’re doing in lab!)
Just coincidence. As a general rule Gram negatives tend to grow faster than Gram positives. We’ll probably isolate mostly Gram negative sin the lab.
Some genera are found all over the place others can show some geographic distribution patterns. There is no hard and fast rule. Some like to quote Baas-Becking: “Everything is everywhere, but the environment selects.”
The bacteria were already there but the addition of oil or NP essentially creates an environment where they have an advantage and become more abundant. We can think of an oil spill as an ecosystem scale enrichment experiment. Also, many of these PAH degradation pathways are carried on plasmids that can be transferred from one bacterium to another (across species, genus, and higher) so some of the isolates could have recently (in an evolutionary sense) acquired the ability to use NP.
NP is often used as a model because it’s safer to use in the lab whereas the 4 & 5 ring PAHs are quite toxic. We’ll see in our next paper that bacteria that can degrade NP can often degrade other PAHs albeit more slowly.
I’ll post the reference section and you can look it up!
I wouldn’t consider it the main question so much as a slightly unexpected result. Both Gram negative and Gram positive bacteria have been shown to degrade NP. It may be that something about their experiment or the area they sampled favored Gram negative bacteria. I mentioned elsewhere that in the BLY 314 lab we typically isolate way more Gram negatives, usually because they grow faster.
Yes there are. Their numbers rose after the DWH spill but are back to normal levels now that the oil is either gone of buried in the sediments or converted to tar balls which are really resistant to biodegradation.
Yes. It catalyses the first step in NP degradation, the addition of both molecules of molecular oxygen across a c-C double bond.
It depends on the chemical properties of the pollutants.
There are some examples of doing this kind of genetic engineering although not usually for PAH degradation as so many “wild” bacteria have the ability. Some enzymes from PAH degradation pathways have been engineered into E.coli to produce indigo and other commercially valuable molecules.
Sometimes variations on catechol are seen but the basic biochemical pathway is very similar across bacterial species.
How do we usually construct phylogenies? look at evolution of genes and genomes? By looking at vertical transmission (parent to offspring). So adding in possible horizontal transmission of genes (between totally unrelated species) will introduce all kinds of confusion into the phylogenies of these genes?
What is the structure of phenanthrene? Does it have more or fewer rings than naphthalene? What did we say about “biodegradability” of PAHs in paper 1?
It could.
Yes and yes. If there’s an oil spill there’s likely to be all kinds of PAHs in the mix.
How would you determine whether a gene has been subject to horizontal gene transfer (HGT)? One way is to compare the phylogeny of the gene of interest (C23O in this case) to the phylogeny of a gene that doesn’y undergo HGT (like the 16S). You’d expect the a gene that undergoes HGT to produce a tree that differs from the 16S tree.
Would you consider the BH medium a defined medium or a complex medium?
Would you connsider the LB medium a defined medium or a complex medium?
How many rings does it have? How do you think this will affect its biodegradability?
Right. You’re doing a scaled-up version of what we did in lab last week, a bunch of tests in a microtiter plate instead of in test tubes. See here for more info and pictures: Biolog .
Look at the numbers in the primer names. They’re not amplifying the entire 16S gene but they’re getting most of it so it will be enough to assign the isolate a genus and most likely a species.
With respect to the database they used, different people have different preferences. The choice doesn’t usually affect the result.
Think about our discussion in class of how to determine bacterial abundance. Why would they dilute the cells to a particular turbidity (=OD) before adding the cells to the test plate.
It is a restriction enzyme. It cuts double stranded DNA at a specific sequence. Restriction enzymes are often used in DNA cloning experiments.
They’re describing the construction of a genomic library here (see https://upload.wikimedia.org/wikipedia/commons/7/7e/Genomic_Library_Construction.png). Basically you cut the ZX4 genome into little pieces and ligate them into a plasmid and put the resulting plasmids into E. coli. Each E. coli gets a different piece of the ZX4 genome. E. coli can’t use PAHs so it’s a good way to find genes that play a part in PAH degradation. You look for E’ coli colonies that have gained the ability to metabolize PAHs or their degradation intermediates (like catechol).
The authors looked for meta-cleavage gens by spraying the colonies with 2,30dihydroxy biphenyl diether. If they were looking for clones that contained a dioxygenase gene, what would they have done?
They’re lysing the cells to release the enzymes into the bulk solution so that they can access the various substrates they’re testing in these enzyme assays.
Think about the citrate test we did in lab last week. All of out lab NP degraders are aerobes and likely possess citric acid cycles but not everyone had a positive citrate test. Why?
Read the comment directly above yours. Submit another comment in place of this one.
You’ll have to wait for the results to get specific information but you can make some guesses at this point. It probably grew well on phenanthrene compared to their other isolates given that they chose it as the focus of their paper.
We’ll find out for sure in the results section but yes, you’re correct.
This qualifies as getting bogged down in details! Our concern is not with how the instrument they used works. We care about what they measured with the instrument and what that tells them about their isolate or clone.
Ask a new question or, if you’re really interested in how FIDs work look it up and share it with us.
Read the previous comment by me for the answer to your first question.
Use google to look up how a centrifuge works. That question counts as getting bogged down in details that aren’t that important to understanding the paper.
There just placing the flasks on a shaker and shaking at 200 rpm.
SalI is a restruction enzyme. It cuts DNA at a specific sequence. If you’re trying to clone a piece of DNA you can cut your plasmid and your insert DNA with a restriction enzyme to generate compatible ends that you can stick together with DNA ligase.
Brittney has it. They’re using the TCM to extract the phenanthrene from the aqueous sample. You probably did something similar at some point in organic lab using a separatory funnel.
exactly.
Because they’re trying to identify phenanthrene degradation genes from ZX4. They made a library of ZX4 genes and put it into E. coli and looking for phenanthrene degradation activity. If E. coli could degrade phenanthrene all their clones would appear positive. They need to use a host that lacks the function they’re trying to find.
The focus in this paper is on describing one isolate in great depth. They probably had other phenanthrene degraders but this was the best or the most unique so they decided to focus on it.
Yes these are fairly standard tests. You can always find more tests to run but given that the focus of this study is phenanthrene degradation by ZX4, so the tests listed are sufficient.
Yes, they use GC. The first sentence of paragraph 3 of the M&M mentions it but it’s really easy to miss. It’s a very standard analysis so many papers will not give the methodological details in the M&M. Here is a link to one of the major companies that do these analyses http://www.midi-inc.com/pages/microbial_id.html
Yes, there are many species of Sphingomonas, so many that in the decade since this paper was written the genus has been split into 2 or 3 new genera!
Many species have been found that degrade all kinds of mono- and polyaromatic hydrocarbons.
There was nothing wrong with them. ZX4 just lacked genes for their degradation or their transport into the cytoplasm.
The number is the measurement of the turbidity or absorbance of the cells and substrate in each well of the microtitre plate. Would you expect a high or a low number if the substrate was consumed by the cells?
They are labeled on the tree. I don’t understand what you’re asking in this question.
Which table? They don’t refer to a table in this paragraph. Try moving your comment to the appropriate paragraph.
It doesn’t produce the enzymes that break down those substances. A similar situation to why we can’t live off cellulose: we don’t have the enzymes necessary.
What do you think? When we looked at pH preferences in lecture what was the “normal” pH range for most microbes?
What do you mean by fix? In biology to fix something is usually to reduce an inorganic molecule into a biomolecule using ATP and NADH/NADPH (ex. carbon fixation and N-fixation).
When we’re looking at phenanthrene (and other PAHs) degradation we’re looking at an oxidative pathway that yields ATP and NADH for the cell. As with any organic substrate it could be respired completely to CO2 or it could be partially oxidized to TCA cycle intermediates which are used in biosynthetic pathways.
It’s possible.
Did anyone compare these concentrations to the naphthalene concentrations tested in our previous paper? This would be a great comparison to include in your summaries of this paper.
We discussed and used this in lab. You should be able to answer this.
No it is not. You need to think about this in terms of how catabolic pathways are regulated. Instead of glucose vs lactose, thin about glucose vs phenanthrene.
Catechol is an intermediate in the phenanthrene degradation pathway. When should the cell make catechol 2,3 dioxygenase?
See my reply to Sarah.
Can you move this comment to the correct paragraph?
It may help to draw the figure 3 gene map out and place the information from this paragraph on it (promoter location, RBS locations) What does this suggest about which genes are transcribed together?
Why would they do this? How would it help them understand phenanthrene degradation by ZX4?
See my earlier comment.
Read on to find out.
Yes. Look at the structures and think about what you learned about the effects of different functional groups on aromatic rings in organic chemistry (an O-Chem II topic, I believe).
There are a few things that can limit growth on PAHs. The first is the toxicity of the PAH. There may be a concentration at which toxic effects overwhelm the ability of the bacterium to degrade the PAH. The second possible limitation is that there is a concentration at which the PAH uptake and degradation enzymes can’t go any faster or biosynthesis of other cellular components can’t keep up so growth stalls.
What other culturing methods did we discuss in lecture?
See my reply to Sarah
Yes it would grow on MacConkey.
I’m not sure what you’re referring to here.
Remember here that they’re just reporting the 16S sequence match. 98% identity means the same species but could be different subspecies or strains. The G+C% and the 16S sequence support each other in categorizing ZX4 as S. paucimobilis.
Yes. They’d need to share some of the features we discussed about the 16S gene in lab Oct. 3rd,
They picked 16S sequences from a variety of previously described PAH degraders. They build the matrix and then plot the data in tree form. The clusters in the tree are dictated by the matric/by the similarity of the sequences used to generate the matrix.
It depends on where the oil comes from. depending on the source crude oil has different percentages of aliphatic, aromatic, resin, and asphaltene type hydrocarbons.
These are two steps on the same overall pathway. In the upper pathway phenanthrene can be degraded all the way to salicylate. The salicylate can be converted to catechol or gentisate which are subsequently oxidized to 2-4 carbon molecules that can enter central metabolism (like TCA cycle) in what’s referred to as the lower pathway.
There are two major versions of the lower pathway: ortho cleavage and meta cleavage. They lead to slightly different end products because of where ring cleavage occurs.
I left out a lot of details but that’s the big picture. The authors are using the fact that ZX4 can utilise and transform certain intermediates to infer which of the possible lower pathways it uses.
CDNB is just a substrate that you can use to assay for GST activity. Some types of GST can use it, others can’t. See this link if you want more information on a GST test kit that Sigma sells https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Bulletin/cs0410bul.pdf.
Exactly!
There are reports of strains that only partially degrade phenanthrene and other PAHs. Sometimes it’s due to mutations in downstream genes that inactivate an enzyme in the degradation pathway. Othertimes a bacterium may not acquire the entire degradation pathway during horizontal gene transfer.
I think you can come up with a few. What qualities would be desirable in a PAH “bioremediator”?
The indigo color from indole does not indicate a gram negative bacterium. Look at your notes for lab 2.
They told you about the meta-cleavage pathway in the results section, reread paragraph 8. It’s a little bit indirect but I think you should be able to figure it out. This is also something you could google quite easily.
They’re just saying that the ZX4 GST is more similar to EPA505 and F199 and less similar to SYK-6 and RW5. The sequence groupings match the CDNB activity. Presumably the differences in the DNA sequences lead to differences in amino acid sequence that lead to this difference in activity. It’s not a crucial finding for the paper but it is interesting.
They didn’t find the full GST sequence or gene. That’s a cloning issue, not a result of mutation. We’ll talk about it in lab tomorrow.
I think it’s a question of breadth vs depth. They’re both valid, thorough studies. They just have different goals: paper 1 wanted to assess/describe the PAH degradation potential of bacteria from the Persian Gulf. This study focuses on one strain and tries to understand how it does it (identify intermediates in the pathway, find the genes that code for degradation).
Do you know how indigo dye is normally produced? It might be that producing it using biotechnology would be more environmentally friendly or cheaper than how it’s currently produced.
Yes.
I think the focus of a lot of these papers and of a lot of bioremediation is to understand which bacteria can bioremediate and how they do it rather than growing up batches or bioremediators and releasing them into the environment. The safety issue is always a concern for regulators.
The location of the promoter, coupled with the distance between phnH & I suggests that they are co-transcribed. The fact that phnG is upstream of the promoter suggests that is is transcribed (and regulated) independently of phnH & I.
Keep in mind also that organisms can use more than one nutritional strategy. One could be both a chemoorganoheterotroph (energy, electrons and C coming from organic sources) AND a photoorganoheterotroph. Many microbes can switch between various C, energy and electron sources depending on the environmental conditions in which they find themselves.
Just about anything that exists in multiple oxiddation states can serve as an electron donor(reduced forms) or electron acceptor (oxidized form).
To answer this you need to look at the nitrogen transformations that occur during nitrification and denitrification. The last sentence may help you out.
Why did your RHD PCR fail? There are a lot of reasons a PCR may not yield any product or the wrong product (an inauthentic product).
It’s hard to compare without more information. For example we’d need to know what the electron donor was when we use AsV as the electron acceptor so that we could guess at the energy yield. We’d need to know what electron acceptor was used in the oxidation of AsIII.
So far so good. We might be able to come up with a few more metabolic strategies for microbes in this system once we read the methods.
I think it’s partly because arsenic is relatively abundant in the Mono lake system. I’m not sure that the other toxic elements you found are equally abundant in Mono lake.
One could imagine using AsIII oxidation to AsV as a detoxification mechanism. The key would be to ensure that there were no microbes present that could convert (reduce) the AsV back to AsIII.
Bangladesh has a huge issue with arsenic in its drinking water.
Exactly. We can often see these kind of cooperative interactions across oxic/anoxic transitions or in environments that switch between oxic and anoxic conditions over time.
Under oxic conditions you’d never use AsV as an electron acceptor, O2 is much better. So if you want to see if AsV can be used as an electron acceptor you need to use anoxic conditions.
We’ll see that some of their experiments were run under both oxic and anoxic conditions but their focus is mostly on the anoxic conditions.
Think about what processes the AsIII and ASV could be used in. Some occur un the presence or absence of O2. Some will only make “energetic sense in one of he two conditions.
I think H2 is also a possiility in this system. It all depends on what electron donars and acceptors are available and their relative Eo values.
The green ponds were dominated by cyanobacteria and presumably oxygenic photosynthesis. My guess is that they were interested in studying other metabolic processes.
No idea!
There are likely some grazers that can dine on the bacteria and archaea in the biofilm.
Mono lake is the the mountains in California. They probably can’t access it between October and April because of snow.
They’re keeping the slurries alive but slowing down their metabolism by storing at 5C. They froze the biofilms to immediately stop all biological activity so that when they extracted DNA and did PCR they would get a snapshot of the microbial community as it was a t the time of sampling.
They’re actually using N2 to create an anoxic atmosphere. It would be better to use argon as it is a noble gas and unreactive but N2 is cheaper so it’s often used instead.
To keep the light out.
With an immunoassay you have an antibody-based assay. In a radio assay you use a radioactive isotope of your analyte of interest as a tracer as it’s often easier to measure radioactivity that particular biological transformations.
For example, when measuring photosynthesis it’s easier to measure the incorporation of 14C-Co2 into biomass than to measure the actual sugars the CO2 was incorporated into. You add the radiolabeled CO2, incubate and then filter the cells out of the solution and measure the radioactivity level of the cells to measure photosynthetic CO2 fixation. The textbook discusses radioassays in chapter 19 (microbial ecology methods).
There are also green sulfur bacteria (anoxygenic phototrophs).
You should really read the previous comments before asking a question. A number of these later questions were already answered.
It was isolated in a previous study by this same research group.
What do the enzymes/proteins these genes code for do? That will answer your question.
It simply means that these sequences are unique. It also suggests that they may not be true arsenite oxidase or arsenate reductase genes.
Statistics.
They were different, that is all. They used BlastX which takes a nucleotide sequence and translates it into all 6 possible reading frames and then compares those orfs to a protein database.
The syringe is N2 flushed so that they don’t add oxygen to the incubatiosn during sampling.
A small amount is taken out an analysed for 73-As or C-14 or whatever analyte they want to measure.
Think about the possible fates of the different radio-labeled substrates they use in this experiment. Why would they want to measure the amount of C-14 on a filter.
It’s your job to figure that out bu looking at the data in the next section!
You can answer this question with a dictionary.
It’s possible but we can’t really say.
These incubations are closed systems. it’s likely that over time waste products are accumulating and growth rates are slowing down. Think about the bacterial growth curve for batch cultures.
AsIII can’t die! It’s just completely removed from the culture (and oxidized to As???).
This is a reasonable assumption based on the data in this graph. The next experiment will test your hypothesis.
I think you have a typo – arsenite can’t be both an electron donor and an acceptor. What are the electrons from arsenite donated to in anoxygenic photosynthesis?
Does the lack of response to shaking (and therefore increased O2 exchange) suggest anything about the use of oxygen in these particular incubations?
What potential processes could be driving these increases/decreases?
They’d like to know “who” is using the AsV and AsIII.
What metabolic process(es) would reduce AsV? Look at figure 4. If AsV is reduced is it serving as an electron donor or an electron acceptor?
Can you rephrase this question? Nothing died but certain analytes were completely consumed. What process consumed them?
I’m not sure. They may tell us in the discussion.
Sounds reasonable to me.
We can’t really answer this “why” question. If you look at the 16S rRNA based tree of life you might conclude that the bacteria are more diverse but they’re less diverse than the Archaea in the red mat biofilm (but maybe not in the water or the green mat biofilm).
See my earlier comment. The lower rates may simply be due to an accumulation of waste products in the incubation bottles.
That’s a reasonable interpretation.
That’s reasonable. The other thing to focus onin this paragraph is how they explain the difference in AsIII oxidation rates in light vs dark incubations, suggesting that in the light both AsIII oxidation and AsV reduction could be occuring leading to the lower overall AsIII oxidation rate in the light.
“After examining this figure it seems that As(III) is the electron donor and As(V) is the electron acceptor. ” you don’t need the figure to tell you this – basic chemistry determines which can serve as an elecctron donor.
The second half of your comment is spot on though.
Right, AsV can be used as an electron acceptor during chemolithotrophic gowth using H2S or H2 as an electron donor/energy source.
The next question to address is what carbon sources the microbes use.
Which bacterium? or are you referring to the biofilm. Remember in these inicubations they used biofilm slurries so there may be more than one type of possible metabolim that can be employed depending on the physical (light/dark) and chemical (added electron donor or acceptor) present in the incubation bottle.
In figure 4 the take home message is that AsV can be used as an electron acceptor during chemolithotrophic growth using H2S or H2 as an electron donor/energy source.
Exactly!
The sulfide can also undergo a bunch or abiotic transformations so we probably can add enough to get complete reduction of the AsV to AsIII. It’s also challenging to get good measurements in the sulfide incubations (look at the errors bars for the sulfide bottles).
They’re testing electron donors. The donors are H2S or H2.
It would suggest that the organisms are not using acetate as a carbon source in the light. They may be using it as an electron donor in anoxygenic photosynthesis however.
Look at the Eo values for both and report back your conclusion.
They would have to make a metagenomic libray and screen it with their aoxB PCR product to find a clone containing the full gene. They could then clone the gene into an expression vector and express the gene to figure out what the protein/enzyme it codes for does.
This would be a great exam question!
Consider some of the methods described in chapter 19. I think there are a few methods you could consider to answer your and Wraychel’s question.
Another good potential exam question.
There’s been work since this paper that showed that one or the arrA genes they reported actually functions as an arsenite oxidase – it preforms the reverse reaction of what the sequence match would suggest. They mention this further up the page in paragraph 7.
They’re for two different processes: arsB is used to pump the arsenic out of the cell – costs the cell ATP of PMF to do this; arrA allows the cell to use AsV as an electron acceptor (conserves/releases energy).
Exactly and the waste product from one metabolic group is substrate for the other group so they cycle the AsV and AsIII continually.
Tee chemistry of the water is that is contains a lot of reduced chemical species that can serve as electron donors for chemolithotrophy. These chemolithotrophic oxidations yield more energy of you have a supply of oxygen to use as electron acceptor. The fact that the ponds are shallow means that some oxygen could potentially diffuse into the water and be used as an electron acceptor to fuel oxidation of things like sulfide using oxygen as the electron accepotor.
“maltonic microbes”?
yes.
The authors suggest that a broader diversity of microbes (with a broader range of temperature optima) carry out the reduction but that oxidation is carried out mainly by Ectothiorhopospira strains due to the match in temperature range of PHS-I and the biofilm oxidation.
It could in theory contribute, but they don’t present any data for reduction by PHS-I so I would assume that they tested for it but didn’t detect any activity. They may state this in the text. I don’t remember.
This is based on the data from figure one and some other data tey don’t include in the paper.
They don’t know “who” is doing the reduction but as long as there isn’t a lot of HGT pr the arrA gene they can use the arrA sequence data to make informed guesses.
The waters are so reducing that there is likely some abiotic reduction of AsV going on but the data show that biological reduction is the main cause of AsV reduction when you supply an electron donor (either sulfide or H2).
With those electron donors they use AsV as their electron acceptor. (no use of AsIII as a donor in this scenario, H2 and H2S are way “better”).
Just in anaerobic b/c it can then be used in methanogenesis.
See figure 4. What electron donors drive AsV reduction?
Which data support this?
Yes.
H2S is abundant in Mono lake so they wanted to test whether the microbes can use it. H2 can be produced during fermentation which could also occur in the biofilms so they tested that as well.
Exactly.
They need to regenerate donor or acceptor, hence the cycle AsIII->AsV->AsIII->AsV etc.
This is a good question. The simple answer is that the genes or enzymes determine it but the real question is why a pathway that’s pretty well conserved to this point produces this variety of intermediates. I haven’t read much on the topic so I can’t answer the question.
The pathways probably aren’t rare. It’s just that people haven’t studied marine systems as much as terrestrial systems.
Most likely. There are a few a few bacteria described in the literature that can only get their carbon from hydrocarbons but most hydrocarbonoclastic bacteria can utilize a variety of carbon sources.
Fluoranthene is a PAH. It’s a class 3 carcinogen and therefore a concern. Like many PAHs it can be produced from incomplete combustion or organic matter.
It’s easier to sample terrestrial environments. For anything other than coastal waters you need a boat or ship! All our isolates from fall semester came from soil I sampled at the base of a telephone pole in front of my neighbours house – almost no effort or resources necessary!
We’ll answer a lot of these questions as we read this paper.
Yes, phn is short for phenanthrene. And yes, bacteria can have “biodegradation” islands in their genomes just as some have pathogenicity islands.
There are likely other bacteria that are closely related to C. indicus P73T but I think this paper may be the first C. indicus genome reported. We’ll see in the methods that they compare this genome to the genome or another Celeribacter strain to identify the PAH-related genes.
Why do you both think it’s a blood agar plate? When I googled “216L agar” all indications were that it is a marine agar.
https://www.google.com/search?q=216L+agar&rlz=1C1GGRV_enUS751US751&oq=216L+agar&aqs=chrome..69i57.559j0j4&sourceid=chrome&ie=UTF-8
The details of the method don’t matter. The key details to get from this paragraph are that they are working with 2 different Celeribacter species and that they isolated genomic DNA from them. How they isolated the DNA is irrelevant – you (the class) don’t know enough yet to assess their choice of methods.
I’ll post the reference section so that you can look up the references if you want more details.
Where did they isolate P73T? Does that influence their choice of growth media?
You need to look up the answers to your last two questions.
Yes, because you want to ensure that the cassette for mutagenizing the target gene only surveives by integrating int the target.
For your first question think back to our discussion of conjugation in lecture and lab last semester.
E. coli was used because it’s genetics are well understood and easily manipulated.
Again, google is your friend for these kinds of question but you don’t really need to know what Solexa sequencing is. What you need to take from this paragraph is that (1) they sequenced the entire genome, (2) they identified all the potential protein coding genes, and (3) they identified genomic islands.
Yes. They use the cre-lox system to knock out a gene they think has a role in PAH degradation and then see how the mutant and wild type behave when incubated with PAH.
They don’t know where the fluoranthene genes are located so they chose a genome sequencing-based approach. What other approaches couold they have used? Think about the second paper we read last semester in 314.
Possibly.
Pay attention to the figure legend. It explains the use of the colors. The first 4 rings show the position of diffecent classes of gene in the genome (color key on the right). Rings one and 4 are hard to see because they don’t have a lot of genes but 2 and 3 show the position of genes from various COG (cluster of orthologous groups).
Ring 4 in clack shows the GC skew. Remember that organisms typically have a relatively even GC % across their entire genome. Areas that deviale significantly were probably acquired by horizontal gene transfer from other organisms.
Good pick-up. Plasmids usually contain non-essential genes. Anything needed for day to day survival will be on the chromosome.
We can’t really answer “why” questions. We can only discuss the implications of the data.
Look this up.
Look this up.
They found a lot of fatty acid synthesis genes, not fatty acids per se. You need multiple genes to synthesize fatty acids. The biochemical pathway have multiple steps depending on what kind of fatty acid you are making. To me 53 genes related to fatty acid metabolism seems reasonable.
You tell me! Make a guess based on the genes they found in them.
You can google this or make a guess based on the content of the paragraph.
The cell won’t dissolve in water! They’re suggesting that P73 produces surfactants that will help it access hydrophobis PAHs etc by solubilizing them in water. They could do BATH or E24 tests like in the first paper we read last semester to confirm this.
I posted some review articles to the SAkai site that you can read for this kind of background information.
The description of the PAH genes they found in this paragraph and elsewhere answers your question.
No. Re-read the paragraph. Those two genes are common sites for the integration of foreign DNA by transposases.
This paragraph tells you that it is a breakdown product of phthalate which is a breakdown product of PAHs.
This is answered in paragraph 27.
Both are possible!
This is a good question. We could calculate the energy released by oxidizing fluoranthene completely to CO2 but the yield to Celeribacter would be much less.
Neither.
What do tRNAs do in the cell?
Some may but probably not all. The Rhizobiales that were the source of teh transferred PAH genes are probably good PAH degraders.
Or it might not carry out gluconeogenesis. It may meet it’s glucose needs through its diet.
You can look this up but for our purposes it’s not important. We want to focus on the PAH-related results.
Right now the only evidence is the sequence homology that lead them to annotate P73_3226 as a benzoate transporter.
They will only knock out the gene they target (0346) but given that many bacterial genes are arranged in operons disrupting one gene will often affect the transcription of genes downstream from it.
They address this in the next paragraph.
Yes. They’re essentially activating the ring. It’s analagous to the ^-carbon phase of glycolysis where you expend 2 ATp to activate glucose before harvesting energy (in the form of ATP and NADH) in the 3-carbon phase.
Yes. There are PCR primers available for some of these dioxygenases so you’ll be able to use a PCR based approach to find some of these genes if we have to wait for the genomes.
Sort off! it’s more that the intermediate would indicate that different genes and pathways are being used.
We won’t use GC-MS as we don’t have one available in the department but we can look for accumulation of colored metabolites and we can also measure growth on various substrates. We can do single gene transcript analysis of our isolates grown on different substrates.
Maybe, but those RHDs could simply be involved in metabolism of aromatic amino acids raher than PAH metabolism.
There are a number of different groups of aromatic dioxygenase (see the tree, fig6?). They’re just saying that previously described fluoranthene dioxygenases were in other branches on the tree.
No, the heavy metals are pollutants as well. They did not use heavy metals. The heavy metals are just as much or more of a problem than the PAHs.
PAHs can be removed when the organisms oxidize them to CO2 when they use them as a carbon source. The metals cannot be removed. They can only be transformed to different oxidation states. We covered this concept in BLY 314 last semester.
That is what they suggest in this paragraph.
They can have a variety of effects. See this Wikipedia entry for more details https://en.wikipedia.org/wiki/Heavy_metals
Sometimes the presence of heavy metals inhibits some biodegraders.
Based on your experience in BLY 314 would it be easier to do a PCR to test for the presence of RHD genes or to isolate the PHE degraders from a site?
What kind of abiotic processes could lead to loss of PHE from the test cultures?
That is the assumption. Catabolic genes should not be transcribed (or should be transcribed at an extremely low level) unless the substrate is present.
It’s free of any other ions that could influence enzyme activity.
It shouldn’t.
The cells need a carbon source to grow and make transcripts. They want to compare transcription during growth on PHE to growth on a control/non-PAH substrate and chose glucose.
The BLY 314 class did the isolation and initial characterization of our strains for us last semester. Our job this semester is to find the PAH degradation genes.
Exactly.
Yes, they will. We’ll also send the P. putida RHD that I cloned out for sequencing to make sure it has no mutations before we move it into an expression vector.
They’re using the technique to amplify up the part of the genome that contains the RHD genes so that they can clone, sequence, and study them. They have primers that target part of the RHD but they need to get the full large and small subunit sequences (and maybe some neighbouring genes) so they’re walking out from the area they have primers for.
They tested it. They just didn’t report the data.
Why do you think they did qPCR?
It’s just a solvent that their analytes of interest will partition to.
Of interest to us is that a lot of sphingomonads capable of degrading PAHs have been described in the literature.
Exactly.
So at lower concentrations the bacterium van degrade all the PHE but at higher concentrations either the PHE level is toxic or the degradation capacity is maxed out and can’t process all the PHE. We saw somethiing similar in our first paper last semester in 314.
The RHD contains iron in it’s active site. No iron = inactive enzyme.
Copper doesn’t affect activity at lower concentrations but at and above 4.03 the PHE removal efficiency declines significantly according to figure S2.
Not sure. My assumption is that both would negatively affect the degradation rate. I’m going to have to read those papers!
Yes, especially because they are present at higher levels in the treatments containing PHE that in the glucose treatments.
As the genes are transcribed and translated the concentration of substrate (and inducer) goes down due to the activity of the transcribed degradation genes. Less inducer present means less transcription = less mRNA.
Yes, copper increasing transcription of the PHE-degradation genes was unexpected. They suggest that it may be because the cells will get more energy to resist the toxic effects of copper if they increase expression of the phe genes.
I think you mistyped, copper (symbol = Cu) promotes high levels of phe-gene transcription.
A previous study found a similar effect for nickel in a Pseudomonas strain.
Caroline has it. The arrowhead indicated the direction of transcription so we can tell which genes may be co-transcribed (and co-regulated). It’s similar to figure 4 in paper 1.
Yes. The reason they did this comparison is that some plants can be used to bioremediate metals from soil and water. They take up the metals into their tissues and you can permanently remove the metal from the soil by harvesting the plants.
Plants can also enhance microbial activity in soils by (1) increasing soil aeration and (2) releasing fixed carbon into the soil to promote microbial growth.
Yes and no. I’ve added a link to figure 1. It shows three possible initial degradation steps described in the text. Two of them are similar to what we’ve seen before: both molecules of O2 added across a C-C double bond. The third possibility is a mono-oxygenation where only one oxygen molecule is added to the fluorene molecule at carbon #9.
Keep in mind that we’re more interested in the genes than in the details of the biochemical pathway. Focus on the big picture which is that there are three possibilities for the first step in the pathway and that two of them look like what we saw in papers one and two.
We’ll see. The sphingomonads often have the upper, lower and meta-cleavage pathway genes located separately, but not always.
Sometimes the initial dioxygenase can attack a wide variety of PAHs but the source bacterium cannot degrade the compound all the way to CO2 because one of the downstream enzymes cannot act on an intermediate.
Yes. The Gram-negatives describes so far have genes that look like they could metabolize fluorene but they can’t grow on it. This could be because they lack a transporter to bring the fluorene int the cell. It could also be a case of sequence similarity misleading us. The true substrate of the described genes may be some other PAH. We should also keep in mind that these initial RHD enzymes often have a relaxed substrate specificity but that the downstream enzymes in the pathway are often more limited in their substrate range.
Also, take a look at the dioxygenase poster outside the door of the lab. You’ll see that Gram-positive and Gram-negative RHDs cluster separately.
Either people haven’t done the studies or they’ve tried to find fluorene degraders but failed. Only positive results get reported in the literature.
Most PAH degradation studies have been done on naphthalene because it is the least toxic PAH (and thus safer to work with in the lab).
They’re working with a gram negative because tat is what grew up when they did an enrichment culture on fluorene. It’s not that they went looking for (or “wanted”) a gram negative, it’s simpy that that is what they found.
See this paper for a discussion of angular versus lateral dioxygenation: https://www.ncbi.nlm.nih.gov/pubmed/12483604. All you really need to know is that they still add two oxygen atoms across a C-C double bond.
If the there are complementary sequences in the primers you can get secondary structures like hairpins or stem loops.
This is correct.
Yes.
The SDS-PAGE allows them to assess whether the cloned proten was made. You usually wouldn’t do it unless you had a problem with the expression experiment (for example no activity against any substrate) and wanted to confirm that the protein was made.
When you put the genes into an expression vector and turn on expression you are over-expressing the protein. The plasmid is a high copy number plasmid. T7 RNA polymerase makes LOADS or mRNA, way more than would be produced normally!
You can’t do the derivitization reaction (which helps with detection on the MS) on a dry substrate. Also, the sample needs to be disolved in liquid to inject int the GC-MS.
They’re just looking at the products formed by the action of FlnA1-FlnA2 on a variety of substrates (only step one of the pathway).
The cytoplasm is an aqueous environment, hence they looked at water soluble products.
They cloned the flnA1A2 genes into a plasmid. They didn’t clone a plasmid. Look at the individual steps described in this paragraph. Break out each step and identify what they did and why?
You need t be more specific with your question. Repeated which experiment or experimental step?
Yes. They want to test the activity of the enzyme the flnA1A2 genes encode. The way to do that is make an expression clone and put it into E. coli (which lacks RHD genes) and measure the activity of the enzyme towrads various substrates.
They were checking that E. coli made the proteins that the genes they cloned into the expression vector encode.
The antibiotics ensure that E. coli maintains the plasmid vector. E. coli is lazy and might get rid of the plasmid if we didn’t maintain selection pressure for the plasmid by growing E. coli on media containing the antibiotic.
This is why we refer to antibiotic resistance genes in cloning vectors as selectable markers. They select for cells that contian/maintain the vector.
The big thing to get out of this paragraph is that there was initially a problem with inactive expression clones so they (1) checked that the proteins were made (they were but the MW was a bit off, which they explain), and (2) changed the growth and induction conditions to increase the proportion of FlnA1-FlnA2 that was in the soluble (and likely active) fraction. Step (2) yielded active protein.
Yes, just focus on the substrate range. No need to get bogged down in the specifics of the biochemistry.
Please explain how they differ.
That is the most likely explanation. The other possibility is that LB126 cannot bring phenanthrene into the cell. It may have a transporter that brings in fluorene but cannot bring in phenanthrene. Both are large enough that they must pass through transport proteins.
We can’t answer that “why” question based on what is presented here and it’s not important for our understanding of the paper. You’d need an enzyme biochemistry course to get the answer!
It means they got no product from LB126 using those primers. Try to move away from the idea of “worked” vs. “didn’t work.” Focus on what the results tell us which in this case is that the “gram-negative” primers did not yield a product which tells us that the primers were not a good match to the dioxygenase we know that LB126 possesses (we know it has a dioxygenase from previoous work looking at the biochemistry of growth of LB126 on fluorene).
No it was not used as a comparison. It is the focus of this paper.
What do you mean by did not work? See my response to Alexis above.
Yes. It’s missing some of the gene and depending on what’s missing it may no longer encode an active transposase enzyme.
Yes, both mean sideways!
They only used two at a time! 15 is the reference number for the primers that yielded the 267bp product.
Think about what you know about the lac operon. Is it ever completely “off?”
I think this comment belong to another paragraph!
What would comsume them? What enzymes are produced by the clone they’re testing?
Read the review article I pointed ye to in the previous section. Remember that I told you not to get bogged down in the details of the biochemistry. Focus on the big picture. Which substrates can the flnA1A2 clone oxidize and which can’t it.
Yes!
For an RT-PCR experiment, yes.
The last sentence suggest that it is banned in the US. This is something you could google and report back to us.
It’s probably the only practical way. Given that it is used in agriculture it would be spread over a large area and it would be impractical to remove contaminated soil and groundwater for treatment.
They tell us what it is in this paragraph.
Probably a bit of both although we have yet to isolate a Sphingomonas strain in BLY 314.
Yes, and a novel pathway (which who knows may be less toxic than previously described pathways).
Yes. Unlike us, they don’t have a colored metabolite to help them but by measuring growth in 96-well plates they can identify mutants. It’s not as easy as looking for loss of indigo production but it does work.
Yes. They isolated the gDNA from the mutants, digested it with an enzyme that doesn’t cut the plasposon (which guarantees that they’ll get the mutated genomic region as well) and then ligate, clone and sequence the plasposon integration site.
This paragraph just gives us the growth conditions for all the experiments. You’ll need to read the descriptions of the experiments below to figure out what controls were used for the different experiments.
I’m not sure what you mean here. PhyML s a tool they used to build a phylogenetic tree. The second sentence lists the comparator strains.
What do we mean when we say a gene is expressed constitutively? How does constitutive expression compare to inducible expression? How would the graph look for an inducible system?
Yes. As Caroline notes, this is differs from the other papers we’ve read and from our assumptions about how catabolic genes are regulated.
Not sure I agree with that conclusion based on what is presented in this paragraph. All tey’re doing here is telling us that based on 25 housekeeping genes, KN65.2 falls within the sphingomonads, Novosphingobium in particular.
It probably has more to do with where the pathway was interrupted and how they screend for mutants. They didn’t just look for loss of function, they also examined mutants with reduced growth. Almost all the mutants could break the aromatic ring (released CO2) which suggests that the mutantiosn were for the most part pretty far down the degradation pathway and not in the initial steps.
The language is a bit imprecise but that’s how I read it and it makes sense given that the insertion is in a transporter which presumably brings carbofuran into the cell for degradation.
The question is how would lack of the transporter turn off transcription and translation of the cfdABCDEFGH genes. It makes sense that you don’t make the proteins unless the substrate is present but how do you think that is controlled (if the system is regulated like lac, what is happening at the promoter and operator and what do you think is the effector molecule?)
I’ve never worked with carbofuran but I believe that you’re correct. The pink color forms when carbofuran is used as a N source. Here is the pubchem page for carbofuran: https://pubchem.ncbi.nlm.nih.gov/compound/carbofuran#section=2D-Structure
The error bars on the graph overlap. We typically take that to mean that the values do not differ statistically.
There is nothing about HGT in this paragraph. They’re just looking at the phylogenetic affiliation (or identity) of KN65.2. They have the genome sequence so they were able to analyse more than just the 16S rRNA gene. They looked at 25 housekeeping proteins which don’t undergo HGT and therefore are useful in determining phylogenetic affiliation.
No purpose. Constitutive expression of a carbofuran catabolic pathway would usually be considered a waste of resources as the species is probably not usually in a carbofuran containing environment.
Figure 2 shows growth on carbofuran. Figure 3 shows either removal (biodegradation) of carbofuran in black circles or mineralisation of carbofuran (where the carbofuran C is oxidized all the way to CO2) in white circles. If the carbofuran is being used as a carbon source we would expect most of it to be oxidised all the way to CO2 (just as glucose that goes through glycolysis and the TCA cycle is). However, the cells might only detoxify carbofuran, where the carbofuran concentration would decrease but we wouldn’t see a stoichiometric formation of CO2. A mutation halfway through the degradation pathway could lead to partial degradation of carbofuran where the black circles go down in value but the white circles don’t increase very much or at all.
They’re all still Novosphingobium. They’re just saying that they had 5 types or classes of mutant (which presumably had mutations at 5 different steps in the carbofuran degradation pathway).
Yes.
No. I read it as being evidence that it could be a HGT acquired gene – it’s only in one other Novosphingobium isolate but it’s also found in an unrelated bacterium (Brevibacterium).
No. Think about scenarios where a mutation in one gene could affect the expression of other genes. One possibility is when you mutate a regulatory gene but there are others . . .
No, the pink color forms when the only C and N source is carbofuran. It’s a result of using the carbofirnas a N source. When a different N source is included in the media the pink color does not form.
Carbofuran is a food source, not a pathway.
No it’s saying that the mutations were in the same genomic region. A contig is a contiguous stretch of overlapping sequencing reads.
They searched the orfs in their genome sequence against a database and looked for homology. The particular tool they used was MaGe (http://13,561.37)
They were also albe to infer the function of some genes by the phenotypes of the plasposon mutants.
This paragraph describes the results of RT-PCR experiments. We did an RT-PCR in lab. Would you consider it natural of artificial?
How do you interpret the results depicted in figure 4? What questions are answered in panel B?
Yes. Because several genes are often transcribed on one mRNA in bacteria a mutation in gene 1 could also knock out genes 2 & 3. Gene 3 might actually initiate the pathway while gene 1 could be for a later step but if you only looked at loss of phenotype you might miss-assign the initial step to the gene 1 product because that is where the plasposon inserted.
All they’re saying in this paragraph is that there is no evidence that the KN65.2 carbofuran degradation genes are plasmid borne. Another bacterium, MS2d, has the genes on a plasmid but that doesn’t appear to be the case for KN65.2.
Because of the possibility of polar effects. See my response to Caroline above.
Yes. It appears to be an indirect effect that is slowing the bug down, probably because of accumulation of a toxic intermediate in methylamine metabolism.
This group of mutants remind us that even when looking at single knockouts we have to consider the organism as a whole.
Yes. The fact that it couldn’t grow on methylamine is interesting given that it had all the methylamine utilization genes. However, this shouldn’t surprise us. It may simply be that it doesn’t have a transporter to bring methylamine into the cell and that the genes are just for metabolism of endogenously produced methylamine.
BLY 314 students should recall the citrate utilization test where some bacteria is citrate negative even through they possesses genes to metabolize citrate. They lack a transporter to bring exogenous citrate into the cell.
This is a good question. They don’t have a specific explanation and just suggest that a toxic metabolite accumulates. It would have to accumulate at a step after CO2 production. Otherwise we wouldn’t see mineralization. It’s not a very satisfactory explanation but would probably require another paper’s worth of research to answer definitively.
Yes, and that in the cfdH mutants only 50% of the ring carbon was converted to CO2.
No. What they said is that the group III mutants produce only 50% of the CO2 that the wild-type does. Mutations in the cfdH gene reduce CO2 formation by 50%.
What do you mean try a different gene? Do you want to do a directed knockout? How would this help you to understand carbofuran degradation? How would you choose your target gene?
Right now, in this paper, we have a pool of mutants that fall into 5 groups and we’re using the genomic location of the mutations, the phenotypes, and the whole genome sequence to puzzle out as much of the degradation pathway as we can.
Yes 🙂
I’m not sure what you mean by no effects or which mutants you’re referring to. All the group V mutants accumulate carbofuran phenol. Figure 6 suggests that formation of carbofuran phenol is the first degradation step. In the next step CfdC attacks carbofuran phenol, adding an oxygen to the furan ring, breaking it. If there is a mutation in cfdC, carbofuran should accumulate.
We’d have to do a qRT-PCR to accurately answer this but probably not. Remember that in Figure 1 carbofuran-induced and uninduced cells had the same CO2 production rates.
They already tested it in figure 1. See my reply to Lisa for how to get a definitive answer.
Yes, when we look at figure 6 (proposed pathway) a number of steps have no enzymes identified, including the initial step(s) that lead to carbofuran phenol formation.
However, the failure to identify that enzyme does not mans that this is a bad paper. It’s an excellent thorough paper that significantly advances our understanding of carbofuran degradation genetics.
Converting the PAHs into TCA cycle intermediates is more efficient that coming up with a completely new metabolic pathway to degrade them. The bacteria just convert then PAh into a compound that can enter a preexisting catabolic pathway.
Why do you think adding inorganic nutrients would enhance PAH degradation?
Good question. You can answer this question with a quick search on PubMed, ScienceDirect or Scopus. Try searching Rhodococcus + bioremediation.
Good question. You should think about the chemical properties of PAHs vs skin (maybe also the physical properties of the epidermis) surface properties for this question.
Think back to organic chemistry. Are tere differences in stability or reactivity between LMW PAHs (like naphthalene) and HMW PAHs with chrysene or benz [a] pyrene? What distinguishes aromatic hydrocarbons from aliphatic hydrocarbons?
This is a reasonable assumption. You could heck out the Rhodococcus microbe wiki page to see if they have any information on this.
We’ll learn later that the ARHD enzyme has an alpha-3,beta-3 structure. Both subunits are necessary for catalysis. The ARHD also requires the ferredoxin and reductase enzymes for catalysis (they are electron carriers).
We used this approach in lab last week. Our R2A plates contained indole which reveals the presence of an ARHD when it is converted to indigo.
We\’ll use it again this week to help identify our PAH degraders.
Which do you think would be harder to degrade all the way to CO2, a HMW PAH (many rings) or a LMW PAH (few rings).
One thing to keep in mind as we read this paper is that when we talk about bioremediation of hydrocarbons like PAHs we’re talking about a process in which the PAH is going to be consumed by the bacterium. It will be oxidised all the way to carbon dioxide or partially oxidized and incorporated into cellular biomass. Either way it will be completely removed from the environment.
The HMW and LMW refers to the PAH, not to the microbe.
PAHs occur naturally in coal, crude oil etc. Transport amd burning of fossil fuels is one major source.
Good question. Maybe you can do a quick search for pathogenic rhodococci.
I think you’ll be able to figure out why they used the spread plate technique after we do it in lab ourselves.
Pay attention to the media used . When they grew strain CMGCZ they grew it in LB , a rich medium. The initial isolation was done on MSM, a defined minimal medium where Fla was the only carbon and energy source so it took the bacteria a lot longer to form visible colonies.
Re-read the paragraph. The bacteria themselves were not subjected to 300C!
What does it mean to wash something? Why do you wash something? Here they are washing the bacterial cells before the next step in their experiment. Why?
You’re on the right track. They grow the cells in medium with Fla. This should induce (switch on) expression of the PAH degradation genes. They them transfer the cells to media with ILCO but first they wash the cells to remove any residual Fla. They do this because any Fla that was carried over to the ILCO incubation could cause an underestimate of the ILCO degradation rate.
bead beating is a way to lyse the bacterial cells when you are purifying genomic DNA. We’ll use a combination of heat and chemical lysis when we extract genomic DNA later this semester.
The first enrichment culture contains 10% soil so that they have a large inoculum to increase their chances of isolating a Fla degrader. After that initial enrichment culture has grown on Fla for a while we assume that the FLA degraders have increased in abundance relative to the non-degraders. A larger dilution can be made in the next step because of the initial enrichment step.
One could also use HPLC or an ELISA-based technique but GC is most common for PAH analysis. It is often coupled with mass spectrometry which gives structural information.
They’re talking about two different genes: the 16S rRNA which can be used to identify a bacterium to genus and (sometimes) species and the ARHD gene which encodes part of the first enzyme in the degradation pathway. These two genes have different evolutionary histories so we wouldn’t necessarily expect that they would exhibit the same sequence homologies.
Look up the structures of all three test PAHs. A priori we’d expect Nap to be most easily degraded, why?
In this case, biology (in particular enzyme structure, ability to bind the substrate) trumps chemistry and even though we might consider Nap the “easier” substrate to degrade, the degradation experiment tells us otherwise.
Look at the graph. 37.8% of the starting Fla was removed by day 3 in the MSM cultures (black squares).
What do you think the yeast extract adds to the medium that may be lacking in the MSM? Probably mostly nitrogen but also other easier to degrade carbon sources.
They want to keep the timescale similar to that in the previous experiment. Why do you think they would want to do that?
Why measure degradation at different concentrations?
How many different hydrocarbons do you think are present in crude oil? Would this affect degradation of ILCO?
There’s a reason we require chemistry. It’s not just to make you miserable!
You can report any % homology. It’s not quite the same as talking about homologous genes and deciding whether they belongf to the same gene family. In this case we know we’re comparing the same gene (16S rRNA) from a number of different species. The usual cut off to consider bacteria members of the same species is that their 16S rRNA gene homology be 97% or greater.
Maybe someone who has taken stats can answer this question? Why perform replicate measurements at all? Why not analyse just one sample for each time point?
To get better Nap or Phe degradation you’d probably need to do your initial enrichment on Nap or Phe.
Compare this graph to the growth curve for bacteria we looked at in class on Thursday. You should see some similarities (especially if you invert it).
The different concentrations let them assess the degradation limits or this Rhodococcus. What’s the highest concentration it can handle, what’s the lowest?
You’re on the right track. PAHs are toxic after all. In theory there should be a concentration at which the bacterium can’t degrade it fast enought o bring the concentration below the toxic level.
You may need a collection of different bacteria, each with a different degradation profile that targets a different fraction of the crude oil.
The Rieske fragment is one of the motifs necessary for the catalytic activity of the ARHA which initiates degradation of PAHs.
We’ll amplify both the 16S rRNA gene and the ARHD gene of your isolates later this semester.
Note that the best matches to the Rhodococcus ARHD gene is from a different genus of bacteria. Did this strike anyone as surprising?
I think some farmers might be upset if there were pesticide degrading bacteria in their fields!
Although, there is quite a bit of work on using bacteria as biocontrol agents, for example Bacillus thuringiensis to kill all kinds of caterpillars.
So if they contain mycolic acids they would stain what color using teh acid fast staining technique?
In this case the clear zone indicates removal of PAH. The PAHs are not water soluble so one way to add them to growth media is to spray them onto the surface of the agar in an organic solvent that will evaporate off leaving a thin film of PAH behind. This looks a bit cloudy and as it is removed a clear zone forms.
probably because it is found in all microbiology labs! Also because it is an easy way to supplement with all 20 essential amino acids and other growth factors as it is just hydrolysed yeast cells.
See my reply to your comment in the results section. There are a couple of potential factors: first is toxicity, second is that transport systems can become saturated.
Something to keep in mind is that bacteria can engage in horizontal gene transfer. Degradation genes are often carried on plasmids which can move between different species (and genera) of bacteria. Just because two isolates are closely related as revealed by their 16S rRNA gene sequence does not mean that they will have similar activities.
Phylogeny does not determine function for bacteria or archaea.
They used a different primer set to produce the 100-bp product. This means the primers bound to a different part of the Rieske motif than the primers that produce the 78-bp product.
The ARHD alpha subunit gene is about 2000-bp long. They only analysed the Rieske center which is only part f the gene.
Two different primer sets were used to target the Rieske center: one set produces a 78-bp product, the other produces a 100 bp product.
You’d need more than the Rieske center. You’d need the entire PAH degradation pathway if you wanted to get PAH degradation.
They can also be generated grilling meat (although in low amounts).
This is true. The background information on PAHs is the same but the focus of the study will differ beyond the fact that they’re studying different genera.
There are a lot of publications on this very topic.
All excellent questions, some of which will be addressed in this study.
They’re interested in petroleum PAHs because the major sourse of PAHs in marine systems is oil spills, whether from blowouts like the DWH or shipping accidents like with the Exxon Valdez.
You can look this up and report back to us! Also, the fur they list here represent a couple of different types of PAH.
I think it’s also cool that a technique we used to identify NP degraders (indigo production) can also be used to identify clones in a genomic library that contain the genes that catalyse the first step in PAH biodegradation (and production of indigo from indole).
Things to look up here. What is an expressin vector? Why colne genes into an expression vector instead of just studying the genes in the clone from the genome library?
Also, pay attention here to the genes they cloned into their expression vector. Why did they clone those particular genes? What proteins/enzymes do they encode?
The only thing you should focus on in this paragraph is what was measured, not how it was measured.
You should be able to find information on Luria-Bertani agar in he Difco manual posted in the resources section of Sakai. It is often abbreviated LB. I’m not sure what else you need to know other than that it is used to cultivate E. coli but the Difco manual should answer your questions.
Review your notes from the biotechnology and genomics sections of BLY 302. They should help you with both your questions.
They’re not interested in the lac operon. They’re using parts of it as tools in their cloning vectors.
Review your notes from the biotechnology section in BLY 302. You should have covered all of this in that class.
I think they give the M9 recipe in a table in chapter 5. It’s an example of a defined medium. Why do you think they would use a defined medium for this experiment?
The effective concentrations have been empirically determioned for “lab rat” E. coli.
Read the genotypes of the plasmids (Km = kanamycin, Ap = ampicillin). Different antibiotics were used to select for different plasmids.
Read on. It should become clear later.
Do they want to study the entire PAH degradation pathway or just part of it?
Think back to BLY 302 and when you learned about Sanger sequencing. How does Sanger sequencing work?
Also, do not ask why particular kits or experimental techniques were used. Ask why the particular experiment was done.
Make sure your comment is attached to the correct paragraph. To answer this question use google! It looks like others have answered the question for you.
Just keep in mind that you don’t have to understand everything about the analytical techniques, you just need to understand why they used them and what information the experiment yielded.
Most journals require that you deposit the sequences in a public database upon publication. It allows others to build on your work (and verify it).
We’ll find out in the results section!
I think we fell down a bit of a rabbit hole here! In any experiment where you are going to do analytical chemistry you want to use the purest chemicals you can because otherwise you might detect a contaminant from your chemicals and erroneously think it was a metabolite produced by your isolate or clone.
We’re actually using it to separate some of the products produced by teh clones from the cells.
Exactly.
What goals did the authors set in the introduction? Are they goals that can be achieved by looking at the whole organism (as in paper 1) or do the require a gene-level study?
In lab we used RNaseA which breaks phosphodiester bonds in RNA between C and U bases. We used it to remove the RNA from our preps.
Restriction enzymes also break phosphodiester bonds but they do it in double stranded DNA and at specific recognition sequences that are 4-10 bp in length.
RNaseA destroys RNA. Restrictionases cleave the genomic DNA into fragments that are a good size for cloning. The LAnder videos discuss this concept well.
The genes will be inserted into the expression plasmids and then E. coli will make the proteins the gens code for and the authors can measure what the proteins do!
What do you mean by “another genetic set?”
The medium is selective if they add an antibiotic to kill all the cells that don’t contain the plasmid. In this case they’re not extracting DNA. They’re going to assay biotransformation of PAHs by E. coli containing cloned genes (on pPhnA) or an empty plasmid. Why did they assay E. coli containing empty plasmid?
I’d say it’s the best substrate.
This is a technical detail you don’t need to worry about. If you’re taking chem this semester see if your chem prof can answer it!
yes.
So they are not concerned with the yield of the product here. They are concerned simply with the structure of the products. But yes, recovery can be an issue in these experiments especially if you don’t know what the product is (and therefore have to guess at the best way to extract it from the culture supernatant).
Yes, the reason we add the indole to our plates is to detect the activity of the aromatic dioxygenase, which suggests that our bacteria can degrade naphthalene or other PAHs.
The authors state in the last sentence that they contain the same fragment 10.5kb fragent of the A5 genome. This often happens when you make a genome library. The videos in the announcement earlier this week show how this can happen.
They will next subclone different parts of the 10.5kb fragment to get the genome region that contains only the genes they want to study. See how parts B and C of Figure one contain different genes from A. They do this because they want to look at some of the genes individually.
You need to read these papers carefully. The details are very important. It can be helpful to draw diagrams of the various clones they describe rather than just relying on the text.
Part A shows the arrangement of the genes in the genome (and on that 10.5kb Sau3A clone fragment of pH1a).
They sequenced that region and used bioinformatic tools to identify the genes and the restriction enzyme sites.
They used some of the restriction sites to cut out the regions in pH1A that contained the genes they were interested in studying.
For example, digesting pH1a with enzymes P and S yields a fragment that contains the genes encoding the large and small subunites of the ARHD (ph1A1a and phnA2). you can go back ti the methods to see exactly how they constructed plasmids B & C or you can look at the map and figure it out looking at the restriction enzyme sites.
Looking at the annotations for orf 6 & 7 in Table 3 they have nothing to do with PAH metabolism. They may have been \”dropped\” in by a transposase at some time in the past. Remember that bacterial genomes are subject to a lot of \”remodeling\” due to HGT.
Look at orfs 6 & 7. They differ in annotated function to the others. How do you think they ended up there?
They’re not hinting. They’re ascribing function based on DNA sequence identity. This is one of the reasons we sequence DNA, because we can use that information to infer fuinction.
Not an anomaly, see my comments above.
Also, we often see adjacent genes reading in different directions. It helps with regulation. Can you explain why?
Remember that we see evidence of things like gene duplication all the time in genomes. It could just be that there was duplication of the lsu but not the ssu. Clearly there is a functional ARHD as A5 and the pH1a clone both produce indigo.
We’re just seeing that evolution is not always “tidy!”.
We’ll fins out below. made expression clones of the ARHD and phnC so that they can figure out their activity.
The level of identity can be an indication of how similar the substrate range of the enzymes are. We might predict that PhnA1 will act more like the protein with which it shares 61% aa identity than the ones with which it shares less that 45% identity.
They’re reporting the % identity to the enzymes listed in the sentence. 51% to the lsu from F199, 62% match to lsu from AFK2.
It’s hard to answer “why” questions in biology. Given that the enzyme encoded by A3 interacts with the A1A2 enzymes we’d expect it to bear a similar level of identity as we saw for those components.
How would PCR accomplish expression of a gene?
What does it mean to express a gene? What is needed to express a gene?
I think you should have covered this in BLY 302 but maybe not. I posted a link to a video earlier this week that should help with your questions.
You can’t publish a paper about the cool new gene you found without learning a little bit about what it can do!
Basically, you need more than just a BLAST match to assign function. You need to do actual experiments to verify the function the sequence match suggests.
What affects the activity level of any enzyme?
How many PAH degrading species are the authors studying in this paper?
You’d typically see greater conservation in a motif such as the Rieske center than in the protein as a whole.
What is the ultimate purpose of the enzymes being characterized in this study?
What is the purpose of the genes they are studying?
I’m not sure what you’re getting at with this question. Location on the DNA strand? Location on a plasmid vs. a chromosome? Your question needs to be more specific.
Why would gene order matter? Probably regulation, right?
They are simply describing that the sequence formed a group with other related sequences on the tree rather than being on a branch all by itself.
Remember that the first step in PAH degradation requires that electrons be supplied to the RHD. The ferredoxin and ferredoxin reductase are the proteins that supply the electrons.
Remember in our intro to metabolisms ferredoxins are a class of electron carrier molecule.
Can you think of an experiment that would answer this question?
It’s just another way of naming or describing the (A)RHD enzyme. The catalytic sites in the enzyme have iron-sulfur clusters (as do many other enzymes).
The relative location of the genes has more to do with the evolutionary history of the horizontal gene transfer events that introduced the genes to the species and any subsequent genome rearrangements/events that may have occurred.
Habitat won’t directly play a role in where the genes are located. It’s more likely that the genes may have been arranged differently in the ancestor of the Rhodococcus in paper 1 and Cycloclasticus in this paper.
What elements in the genome are involved in regulation of gene expression and how would location play into regulation (if at all)?
They’re just saying the arrangement and location in A5 are unique compared to what was found in previous studies.
When these PAH degradation genes were first described they were all on plasmids but if you think about what you know about bacterial genomes from BLY 302 there is no reason some of these genes wouldn’t be found in bacterial chromosomes due to transposition events.
The phnA1 gene encodes an RHD-alpha subunit. It doesn’t use any proteins. The branching pattern on the tree tells us which other RHD-alphas it is most closely related to. The fact that it falls outside means that while it’s a true RHDalpha, it differes from the majority of previously described RHDs.
Try not to focus too much on the numbers. We don’t know enough yet to compare 44% identity to say, 60% identity in a meaningful way.
Probably just transposases from transposons/insertion sequences. CRISPR in its “natural” setting is for viral defence, not moving genes around.
The substrate range of the enzyme is just a characteristic of the enzyme. It doesn’t affect the enzyme itself. It does affect the organism that contains that gene/enzyme, right? It affects what the organism (Cycloclasticus A5 in this case) can use as a substrate for growth and energy production.
Maybe, maybe not. I can’t remember if they told us if A5 can grow on anthracene in the intro or M&M. Growth on anthracene would suggest that another enzyme is present in A5 that initiates degradation of anthracene given that PhnA cannot.
Can anyone describe what is meant by upper and lower pathways? I think this is key to understanding this section. Is the pathway I pasted at the top of the page an upper pathway, a lower pathway or both?
Probably METAtranscriptomics as we’d be looking at the entire community but yes it would be a great tool here especially as the tell us in the next paragraph that they couldn’t amplify true arsenite oxidase genes with their arsenite oxidase PCR primers.
Yes. Some communtiy members are using arsenite as an electron donor during energy generation. Others are using the arsenate generated from arsenite oxudation as an electron acceptor during their metabolism and generating the arsenite. The arsenic is being cycled from AsIII to AsV to AsIII to AsV etc. etc.
It’s the same story that we see with NAD+ and NADH in the cell. NAD+ is reduced to NADH during oxidation of glucose (in glycolysis and TCA cycle) and the NAD+ is regererated when NADH gives up the electrons to the ETC.
Cells are the original reusers/recyclers!
Great work seeing the parallel with our pahE PCR “failure” in lab. Sometimes primers that work for some organisms don’t work with the bugs you are studying.
Thank about other strategies they could use to find the arsenite oxidase genes. Sarah Grace had a good suggestionin teh previous paragraph.
It’s possible that one form is more toxic than the other. Look at arsenic’s position in the periodic table. Any ideas?
All will be revealed in chapter 14! For now think about whether sulfide, ammonia, or methane could serve as electron donors (and thus energy sources).
So they are going to do two types of analysis on the biofilms.
1) DNA analysis – to see “who” is there and “what” they can do.
2) Incubations – add the biofilm to media and measure the various transformations (of AsIII, AsV, various carbon and electron sources) the biofilms carry out.
Do you think the biofilms will be metabolically active if you store them in the dark at 5C?
Why do you put your food in the fridge? Maybe they have a similar reason for putting the biofilms in the fridge before they start their experiments.
The samples are essentially the same, the experimental treatment varies.
They describe a number of different experiments here. Some look at As transformations. Others look at C-use. It will make more sense when you see the data in the next section.
In the first experiment they are looking at arsenic transformations. The key thing to know here is that radioactive As is added in very small amounts. It is a tracer. Most of the As in the experiment is unlabeled.
Remember that in the introduction they told us that these biofilms show evidence of respiratory As(V) reduction, photosynthetic anaerobic As(III) oxidation, and aerobic As(III) oxidation. This section describes the incubation experiments that lead to those insights.
Journals require that authors group similar experiments in the same section. Part of your job as a reader is to separate them out again. Take notes as you read this paragraph. Pay attentions to electron donor and electron acceptor as well as carbon and energy source.
They have two ways to assess whats happening in the biofilms:
1) determine what genes are present
2) measure various biochemical transformations (e.g. AsV -> AsIII.
This paragraph describes #2.
Presumably reference 4 describes that work. I’ll post the reference list so you can look it up (the title will probably let us know).
The physical and chemical description of the site gives us the information we need to form hypotheses about what kind of metabolisms might be found there.
Hang has it. They’re preserving the samples in an inactive state so that they can do their experiments later with essentially unaltered samples that represent the state the biofilms were in when they originally sampled.
We don’t care about evolution here. We want to determine what the bacteria/archaea can do with the various C, E, and e- sources – how they use them in their metabolism.
What metabolism requires light and can’t occur in the dark?
Think about available electron acceptors.
They pool three independent replicates in case the PCRs behaved differently. It should give them a more representative sample of the community in the biofilm than if they just did one PCR.
With only 96 total clones analysed they discarded the singlets because they’re not confident how representative they are. They just focused on the more abundant groups. It’s not critical for our understanding of the paper.
Think about what they will do with the samples after storage. For incubations they store at 4-5C (slows things doown but doesn’t kill the cells. For DNA analyses they store at -80 – kills the cells but preserves the DNA.
They want to wash away any e- donor from the sample and put it in a medium that contains precise amounts of the donor they want to study.
Remember that when we discussed genome sequencing we talked about how a certain % of the orfs identified in a genome either have no match in GenBank or the sequence they match has no annotated function.
With degenerate PCR primers you often amplify non-target sequences and many of these have no match in GenBank.
They include killed controls in order to confirm that any transformations they see are a result of microbial activity and not spontaneous chemical reactions.
What’s interesting too is that the Archaeal community seems to be more diverse and more even than the bacterial community.
The amount of light is constant. In the light the AsII goes down and AsV gos up, so AsIII is being oxidized to AsV. What possible process(es) could be using the electrons removed from the AsIII. Make a list!
As you look at this graph think about what process(es) could take AsV and reduce it to AsIII in the dark. AsV -> AsIII is a reduction. The AsV is being used as an electron acceptor by what possible processes?
Lastly keep inmind that these incubations were carried out in sealed bottles (i.e. a batch culture). Over time waste products accumulate in sealed systems and can “slow” down microbial metabolism.
Exactly!
Remember that it’s not just one organism. it’s a community. They can coexist by using different electron donors and acceptors in their metabolism and by supplying engaging in syntrophic interactions where one organism’s waste is another’s fuel.
Yes. if they didn’t recycle and couple the AsIII oxidation with AsV reduction they’d run out of electron donor and acceptor rather quickly.
Addressing both Erica and Christy’s question. One narrow optimum temperature for oxidation might suggest that it’s only one organism that is carrying out this process. The broad temperature range for reduction could be because a variety of different bugs are reducing AsV (each with a different temperature optimum).
The PCR and sequencing of the reduction and oxidation genes could help answer this. if there is just one reduction gene sequence it would suggest just one bug does the reduction. If there are a variety of reduction genes, many bugs reduce AsV.
Yes. Other studies have shown this (references 7 7 20 which they cited).
It’s “good” in that it’s always nice when a follow-up study confirms the findings of a previous study.
Also, as we discussed in class, isolates are not always ecologically relevant. In this case the culture independent 16S rRNA analysis confirms that their previous isolate is in fact a major player in these biofilms.
While possible, these basic metabolic genes don’t usually move by HGT.
Remember that these biofilms probably straddle the oxic-anoxic interface in Mono lake so many of the microbes can make a living under both O2 regimes.
Compare it to the time scales in the other experiments. Is it that different?
What do you mean by very similar? They\’re mirror images of one another. In one AsV decreases and in the other AsIII increases.
Why does this happen? The AsV is reduced to AsIII, so AsV goes down and AsIII goes up.
The critical thin to identify here is where the electrons used to reduce AsV to AsIII come from. What else decreases in the top panel of the graph?
At the time this paper was written arsenite oxidase was not well studied so their primer options were limited.
definitely, or transcriptomics.
Not quite sure what you’re getting at here. Remember that 16S genes are not usually subjected to HGT. The finding of only one type of bacterium in the biofilm simply suggests that the biofilm is essentially a monoculture on the bacterial side.
HGT might play a role in arsenate reduction but may not. remember that this ability evolved early in life in earth and would be expected to be found in many phyla (at least those still living in places like Mono lake).
Can any of you think of an experimental technique from chapter 19 that could answer this question?
This would be a great exam question!
Mono lake experiences temperature extremes. In summer the temperature can range from 6 to 36C. It’s high in the Sierra’s and gets >60 inches of snow a year.
Until they do the experiment all they have is a hypothesis. They had to do the experiment to gather evidence to support or reject the hypothesis that H2 and or Sulfide oxidation was a major component of the lake biology.
But there could be oxidation of acetate in combination with reduction of a different electron acceptor… The key is the second sentence of the next paragraph…
They’re saying that acetate is not oxidized in conjunction with AsV reduction. In the next paragraph they tell us that acetate is oxidized under a different metabolic scenario.
read this paragraph!
Many Archaea are chemolithotrophs. They may be maing a living by either sulfide or hydrogen oxidation.
They subsequently found (after doing some genomics) that the arsenite oxidase in this system \”looks\” like an arsenate reductase sequence-wise. It has some key substitutions that make it work in reverse!
It’d be surprising to find activity at temperatures significantly higher than the biofilms experience in situ.
Equal and opposite reduction of what? They saw acetate oxidation in the light but not in the dark. Where do the electrons from acetate end up in the light?
So in photoheterotrphy the energy comes from light (photo) but the C comes from organic molecules (hetero). A bit confusing because we often think of heterotrophs meeting their C, e-, and E needs from an organic molecule bur microbes are very metabolically versatile.
Hopefully this drives home that it’s not just photosynthetic organisms that fix CO2 into biomolecules. Chemolithoautotrophs do it as well. Some use the Calvin cycle but others use some of the other carbon fixation pathways we touched upon in lecture.
Sometimes bioremediation occurs without any manipulation of environmental conditions. for example in the aftermath of the DWH blowout in 2010 microbes in the GoM degraded a lot of the released oil without any special measures being taken.
Rates of bioremediation are quite variable and depend on the exact substance being degraded, its concentration and other environmental conditions (think about things like temperature and availability o essential nutrients such as n, P, K and Fe which could limit microbial growth).
How does temperature affect biological processes? chemical reactions? Does it have any effect?
Remember, focus on the big picture for each experiment, not the details.
No control need for collecting the sediment as this is a discovery type study. They’ll run controls later though when they’re measuring PAH degradation etc.
What stress do you think they’re referring to here/what stressful condition are they exposing the bacteria to?
Read on to find out!
They’re doing an enrichment culture, like Beijerink in chapter 1 and like we’ll do in lab next week. Read carefully – they add the PAH as a sole source of carbon so only PAH metabolisers will be able to grow and reproduce.
The glycerol stock at -20 are for long term storage.
They’re not comparing the isolates to E. coli. The numbers in the primer names (27 and 1492) refer to the position of the primers in the sequence of the E. coli 16S rRNA gene.
Review the section on electron microscopy in chapter 1 for the answer to this question!
What does pH measure? What does conductivity measure? Why would these affet living organisms? Think about the basics you learned in oter classes about proteins and enzymes.
There’s wave action over the top layers of sediments in the ocean. Also, various animals (worms etc0 disturb the sediment allowing oxygen exchange into the sediments. by shaking at 100rom they mimic this. They need this gas exchange because they’re looking at aerobic PAH degradation.
One reason is the size of the gene (~1500bp). It’s easily amplified by PCR and gives sufficient information for good phylogenetic discrimination. The 5S rRNA is only ~120bp and the 23S is ~3000 bp.
It allows them to graphically depict the relationship of their isolates to the entire diversity of the bacteria. It can be more valuable than simply looking at the information the BLAST search gives you (% identity to closest relative). For example, it’s easier to see on a tree if all yor isolates are closely related to one another.
Yes, they’re making a concentrated solution of bacteria. Why do you think they did this before adding the PAH?
Think back to organic chemistry and to freshman biology. What kind of solvent is acetone? What would dissolve in it?
Read my introduction to this section. Focus on the big picture – yo don’t need to understand all the little details, just why they did each experiments. Most comments on this section got way to bogged down in technical details.
Do you really need to know the details of the Kimura method beyond what is in this paragraph? If so, look it up and report back to us.
Take a look at the map (figure 1 in the previous section). It may help explain the physico-chemical differences between sites. Keep in mind that conductivity is related to salinity. The key thing in this paragraph is that they sampled 3 different sites – all the sites had detectable PAH (so they would reasonably expect to find PAH degraders) but the sites had different nutrient profiles which could limit bacterial productivity (PAH can supply C and energy but they need N, P, K etc. to build proteins, nucleic acids etc.).
This is a good observation. An isolate with a wide substrate range would be more “useful” than one with a narrow substrate range for bioremediation as crude oil is a complex mixture of hydrocarbons.
Keep in mind that in nature we have a community of microbes with different abilities carrying out bioremediation. We don’t rely on just one “superbug” to do all the bioremediation work where PAHs and crude oil are concerned.
Most often we rely on bacteria that are already present and either add nutrients (N, P, K etc) that may be limiting their metabolism or add a supplemental carbon source that allows them to grow faster and co-metabolise the PAH (but not rely on only the PAH for growth which would be slower).
That could be true. it could also be that the media they used in lab is not the best choice for growing these bacteria – we discussed this in lecture when we talked about potential explanations for the great plate count anomaly.
You have to do microscopy to determine the morphology of the cells.
Light microscopy will do. You’ll do this with your isolates in week 4. No SEM for us in BLY 314 but it’s not necessary for determining rods vs cocci.
Gram status shouldn’t make a difference. Gram negatives are often faster growing (probably just an artifact of the media we use in labs) but it doesn’t mean that they will degrade PAHs faster. There is some evidence to suggest that Gram positives like Mycobacteria are better at degrading HMW PAHs.
If you’re referring to panel 5 it looks like a capsule surrounds the cells in that image.
We use cocci and bacilli to describe cell morphology, not colony morphology. For panels 1-2 of figure 2 you’d use the terminology in the lab 1 protocol and describe colony shape, margin, elevation etc.
Based on 16S rRNA gene homologywe’d classify them as the same secies (rule of thumb is >97% identity is the same species). However, we need to keep in mind that they didn’t sequence the entire 16S gene, just part of it.
Also, the 16s gene is just one marker. It’s not perfet and 100% 16S rRNA sequence identity doesn’t mean that the two strains are identical throughout the genome, especially as bacteria can engage in horizontal gene transfer via conjugation/ transduction/ transformation and “pick up” genes from their environment.
Doing a partial sequence was probably partly a budget issue. Science costs money! Sometimes you don’t have enough to do a perfect experiment but you can still generate good, publication quality data.
We’ll do partial 16s sequences for our isolates this semester. They’ll allow us to identify our bacteria to at least genus level and probably species level.
Do you know what the KEGG model is? It’s the Kyoto encyclopedia of genes and genomes. It’s useful for genomic and biochemical studies. Why do you think they’re using it here?
This is a good question. Bacteria are more complicated to analyse that plants or animals as function does not always follow phylogeny (because of HGT). However, identity and relatedness is still a good place to start.
So, for Ochrobactrum there’s some previous work showing this geneus can degrade PAHs. Their work asdds to our understanding of the role of Ochrobactrum in degrading PAHs.
The Cupriavidus result may be the more interesting as they tell us not a lot has been published on PAH degradation by this strain. This art of the paper represents “new” knowledge.
A clear example of how a picture can be worth 1000 words. Note how for LC there is no difference in branch length (horizontal axis0 between it and the other species on that branch – they’re identical.
However, isolate LA is on a much longer branch from it’s closest relative, indicating it’s not as closely related.
It’s been isolated from a lot of cystic fibrosis lungs. Genomic studies have shown that some Pa isolates have a lot of pathogenicity factors. However, we’d want a lot more data that just a 16S sequence to conclude that their LC isolate is a human pathogen.
Maybe they’ll do a follow-up paper focusing on this isolate.
Remember too, they got LC from a contaminated environment. It’s part of the natural microbial community there.
There’s a small rise at the end of the graph, yes, but we needn’t be concerned with it. We’re just interested in the 15 minute peak for (a0 as that is where phenanthrene elutes and the 28 minute peak for (b) as that is where pyrene elutes. Everything else is irrelevant.
A good question that I’ll leave for all of you to ponder. What confers the ability to do anything on am organism?
I’d encourage all f you to draw the structures of these compounds and revisit your organic chemistry texts and think about the properties of aromatic compounds.
A report is a previous publication an there is no previous publication reporting that Pc degrades phenanthrene. there are previous reports that it degrades other substances but not phenanthrene. this is the first report of it degrading phenanthrene.
PAHs are not totally water soluble. They form tiny particles in an aqueous solution like growth media. this means they increase OD in the beginning (before the bacteria start to degrade them) but as they are degraded they contribute less to OD so the PAH contribution to OD decreases as time goes on.
Naphthalene is the simplest PAH – only two rings. The larger PAHs with more rings tend to be much more toxic and more difficult to degrade. However, the initial step in aerobic degradation of all PAHs, addition of both molecules of molecular oxygen across a C-C double bond, is the same.
Think about differences between deep-sea and shallow water environments. Would you expect coastal (shallow water) bacteria to thrive in the deep? Why or why not? Temperature is a factor but there\’s another big difference that you\’re missing.
You could click on the Pearson et al link and see what that paper has to say about the topic. It migth help or it might be too technical for us in BLY 314. At most I\’d read the abstract and introduction.
Read the first sentence of the paragraph really carefully. It is the key! The technique is also described in our textbook. we won’t cover it for a while but you can look ahead.
Is the overall goal of the study to degrade PAHs? I don’t think so. They want to study the diversity of the hydrocarbon-degrading bacteria at Guaymas. How will SIP help them do that?
Sort off, in that the hydrocarbons originate in the deep subsurface and are moving up into the deep waters of the ocean. However, the physical conditions, i.e. temperature, are quite different which probably has an effect on both hydrocarbon composition and microbial community composition.
I think this is a plausible explanation based on what we’ve read.
Think about what happens when super hot (200degreees +) percolates up through the sea floor into water that is 4-5 degrees celcius).
Temperature can act as a selective factor – favouring growth of some organisms and inhibiting others. The vent fluids may also affect what grows where given that they have a different chemical composition than “normal” seawater.
They’re basically doing a survey so they grab samples from a variety of different locations to see what’s there. A control sample would be at some distance from the vent where there was no vent effect.
It’s a sediment sample where they collect a tube of sediment that spans the surface and down through carious depths.
1. They perform killed controls so that they can attribute any changes to biological activity, not spontaneous chemical reactions.
2. Triplicate means 3 independent phenanthrene incubations, 3 independent ANT incubations etc., etc.
Exactly. As little as 15% glycerol acts as an antifreeze to protect the cells when you store them at -80C.
Adding the PAH makes the media cloudy. If the bacteria metabolize (and remove) the PAH the media turn clear. We’ll do something similar in lab 5 when we look at metabolism of starch and of milk proteins.
We’ll do 16S rRNA PCR and sequencing of your isolates in a few weeks.
So using isotopes (both stable and radio isotopes) allow us to tell if an organisms uses a substrate labeled with that isotope.
In both cases the labeled carbon has two possible fates – it could be partially oxidised and incroporated into cellular biomass or in could be completely oxidised to CO2.
The PAHs are apparently more likely to stick to the glass than to the foil.
So the key here is that they want to separate the DNA from cells that incorporated the 13C-labeled PAH from the DNA of the cells that did not incorporate the labeled PAH. The labeled DNA will be heavier because it contains C-13 instead of C-12.
The E.coli DNA is unlabeled and serves as a standard for unlabeled DNA. It should migrate to the same spot in the tube as the DNA from the unlabeled sediment bacteria.
They can use 16S PCR and sequencing to verify that they properly separated the C-12 and C-13 DNA because there should be no E. coli in the C-13 DNA because E. coli doesn’t live in vent sediments.
The takeaway from this paragraph is that they amplified, cloned, and sequenced the 16S rRNA from the heavy DNA to identify the PAH degraders.
You need a certain amount of DNA for the technique to work reliably and reproducibly. That’s probably why they skipped the low DNA samples.
You would think so but for the most part the pressure change doesn’t seem to affect the microbes isolated from these types of sample.
They describe the differences in the previous paragraph.
Why do you think they performed qPCR instead of regular PCR?
Presumably they got PHE degradation in their earlier incubations and decided to try to isolate PAH degraders from PHE enrichment cultures. Also, the fact that it was one of the PAHs they detected in situ made it a good choice.
People often use naphthalene for these experiments so may they wanted something a little different to see if they would get different bacteria.
They first grow them in pyruvate to get a lot of biomass. They wash the cells to remove any residual pyruvate so that when they add the 14C-PHE the cells are “forecd” to use it because all the pyruvate would be removed.
Microbes will always want to use the easy substrate (gulcose before lactose and pyruvate before phenanthrene or any other PAH).
Sequencing of ribosomal RNA is a common tool to identify/categorise organisms (16S for bacteria and archaea) and 18S for eukaryotes. Organelle genes (for example cytochrome oxidase) are also used in studies of eukaryotes.
For bacteria, 16S rRNA sequences that share 97% identity or higher are considered the same species. We’ll spend teh next 3 weeks in lab working on generating 16S rRNA sequences for our NP isolates.
Find your notes on PCR from BLY 302!
You can find out by seaching the genera in PubMed or another scientific database. As far as I know Cycloclasticus has only been reported from marine environments. I think Halomonas has also been found in saline terrestrial environments as well as marine environments.
Draw it out. I’ve done it for you here but you should all get in the habit of doing this if the text is confusing you. I imagine Madelyn is not the only one confused by this section!
The bacteria don’t use the sulfide instead of oxygen. They use sulfate (oxidised) as an electron acceptor instead of oxygen and produce sulfide.
The last sentence tells us why they focused on PHE in their experiments – because it was the PAH that was mineralised (oxidised to CO2) to the greatest extent by the 4567-24 sediment sample.
Yes, this graph makes clear that they should focus on the 4567-24 sample (open bars) and use PHE as their model PAH because it shows the greatest mineralisation.
Yes. They used the C-12 and C-14 incubations to track the degradation or the C13 PAH. They want to isolate the C-13 labeled DNA from the bacteria that were the first users of teh C13-PAH. They don’t want the label distributed across multiple trophic levels.
They also don’t want to “waste” their C-13 labeled sample so they use the other 2 incubations to track PAH uptake and mineralisation.
They want to look at the microbes that do the initial uptake of PAH. They don’t want to allow any trophic transfers.
Keep in mind that they are comparing killed controls (where they expect no PAH removal/CO2 generation) with live experiments (where they expect both PAH removal and CO2 generation). The circles measure one thing the squares another. A good exercise is to predict what the graph should look like before you look at it.
For the circles they’re measuring the accumulation of CO2 from mineralisation (oxidation/degradation) or the PAH. In the live incubations this line should go up over time and stay flat (and near zero b/c there are no bugs to degrade the PAH) for the killed control.
For the squares they’re measuring the PAH directly. For the live incubations this line should go down over time if degradation occurs. The killed control should be flat but high (ie the amount of PAH added initially).
Probably not as it would likely diffuse into the surrounding sediment and seawater.
Sometimes we see a similar yellow metabolite accumulating in our degrader plates in BLY 314 but I haven’t seen any this year.
So they centrifuge the DNA in order to separate the heavy C13 DNA (from the phenanthrene degraders) from the light C12 DNA of teh non-degraders (and their E. coli control).
Each lane in the gel represents a different layer (fraction) from the CsCl gradient. Notice that the fraction son the left contain different bands than the fractions on the right and that the middle fractions are a mixture. This (and the presence of the E. coli band in only the right-hand fractions tell us that they did a good job separating the C13 DNA from the C12 DNA.
They could have highly similar 16S rRNA genes but differ at other loci in the genome that would allow them to occupy different niches in the ocean.
They’re saying their culture work supported the SIP work. They found Cycloclasticus 16S sequences with SIP and they were able to isolate Cycloclasticus from their enrichment cultures.
I think this is also an encouraging result in that naturally occurring bacteria exist that can naturally remediate these pollutants. Also, the system is resilient – many differnt genera seem to be able to degrade PAHs. We don’t need to engineer any superbugs!
As Abrianna reported, neighbour joining is very fast so it’s very popular. We’ll use it and a few other algorithms in lab once we get teh 16s rRNA sequences of our isolates.
I read this as they can only gow on PAHs if they’re obligate degraders whereas the non-obligate degraders can grow on a wide variety of carbon sources.
They grow slowly, much slower than our bugs in lab on R2A or even MSM!
Yes, they didn’t directly show PAH degradation by Cycloclasticus. The showed PAH carbon in Cycloclasticus DNA using SIP but another organism could have metabolised the PAH and Cycloclasticus could have consumed that organism. This is why they spent a lot of time figuring when to isolate the DNA from the SIP experiment.
The definitive experiment would be to incubate their Cycloclasticus isolate with PAH as teh sole C and E source and measure PAH removal, mineralisation and incorporation into Cycloclasticus.
Yes.
I think it’s helpful to draw the expected results before you look at them. It forces you to think through the experiment.
Remember in BLY 302 you learned that bacteria can acquire genes from many sources. They undergo horizontal gene transfer (from other bacteria, viruses, etc) as well as vertical (from parent) so bacteria can be very closely related as determined by 16S rRNA phylogenies but have different phenotypes (due to the genes they pick up via horizontal gene transfer.
Not location. i used loci in the genetic sense of meaning a chromosomal segment or a plasmid that is present in one strain but not in another.
Probably non-obligate (i.e. facultative) from the bacterium’s perspective as it would mean that the bacterium is metabolically flexible.
A singleton is a sequence that occurs only once. They are often discarded from sequencing datasets for statistical reasons.
Definitely – for the summary you want to distill the information down to the essentials.
A good question. We’re often most interested in the toxic effects of substances and overlook (at least initially) that there can also be non-toxic uses of them (or useful applications of their toxicity in things like pest control).
Yes and elemental selenium is a solid so it’s not as bioavailable as the water soluble selenate and selenite. We can think of this system as a win-win – the bacteria can survive without O2 by using, for example, selenate, as an alternate electron acceptor and detoxifying the environment at the same time.
Yes, it’s pretty cool!
The earth is an oxidizing environment (exposed iron rusts etc) so it could be that abiotic processes re-oxidize the reduced selenium. It could also be that there are bacteria out there that can use reduced selenium species as electron donors (get their electrons and energy from oxidation of reduced forms of selenium instead of from preformed organic molecules like glucose).
Yes but with one key difference. In bioremediation of organic molecules like PAHs they can be completely removed from the environment by oxidation all the way to CO2. In biomineralization the element remains in the environment but it is immobilized and much less bioavailable because it is now and insoluble solid mineral.
Look at the reaction they’re looking for. You should be able to tell whether it is more likely to occur with or without oxygen based on how the cell is using the selenium.
I’m not sure what you mean by this question. Can you rephrase it?
Excellent observation. They definitely won’t be able to use K-12 as a host for their clones or as a negative control but there may be other strains of E. coli than don’t reduce selenate that they’ll be able to use.
Gram statues isn’t closely tied to metabolism.
This is a general methods paragraph that describes the various media they used. We’ll find details of when the media were used in the subsequent sections. Sometimes they’ll tell us explicitly, other times we’ll have to figure it out from this paragraph and the information in table 1.
It may also have been done to make the chemical analysis easier. LB medium is a very messy background to try to measure selenium species in.
I would say that takeaway one is they made a genomic library using a cosmid vector. #2 is they screened the library for selenate reductase activity.
Read the part about the strains again – they’re testing the E. coli hosts, not the cosmid clones.
They wanted to screen the library for clones that contained DNA that conferred the ability to reduce selenate onto the E. coli host. Therefore, they first screened various E. coli hosts for the ability to reduce selenate. They needed a selenate reductiion-negative host.
Review section 12.4 in the textbook (cassette mutagenesis) for help with this section. You don’t really need to understand the details, just that they are trying to knock the fnr gene out of the wild type E. cloacae.
Think about what the cells are using oxygen for and what they would use instead of oxygen once the oxygen ran out.
This is not what I want you to focus on. You are getting bogged down in the technical details! Focus on the big picture. We don’t care how they isolated the DNA we care why they isolated it – what did they do with it and why did they do it?
See figure 12.8 in the text or time travel back to the pGLO lab in BLY 121 if you took the 121 lab here. What would happen if you didn’t include the antibiotic that selects for the cosmid in the agar?
How would they tell if they had a clone that encoded selenate reduction if they put the cloned DNA in a host that is positive for selenate reduction?
It may help to review the section on cassette mutagenesis in the text and to cartoon out the experiment. The knockouts will only carry one of the antibiotic resistance genes from the sacB based vector (The sacB-based vector suicides when you add sucrose to the growth media).
Their goal is to find genes involved in selenate reduction. Based on their knocking out fnr and then complementing fnr it seems that fnr has something to do with selenate reduction by E. cloacae.
We don’t care about the technical details! Focus on the question, not on the technical details.
Their clone produces a product so they analyse the product.
Remember there is no degradation of Se, there is only transformation from one form to another. The Se is never removed/degraded the way an organic compound can be removed by oxidation to CO2.
There is no degradation of selenium there is only transformation from one form to another.
Cosmids can accept larger DNA fragments than plasmids so when you make a genomic library you don’t need as many clones as each clone contains a larger piece of DNA.
You need to pay attention to the names here. E. coli S17-1 does NOT reduce selenate. They told us this in the methods. The lack of selenate reduction is why they were able to use it as a host to screen clones for genes that confer the ability to reduce selenate.
Also, S17-1 plus the empty cosmid vector (pPLAFR3) does NOT reduce selenate.
However, the red colony/clone that contained the cosmid pECL1e does reduce selenate. The conclusion is that one of the genes inserted into the pLAFR3 cosmid to yield the pECL1e cosmid confers the ability to reduce selenate on the E. coli S17-1 host.
No, the graph shows that the pLAFR3 clone does not reduce selenate at all. The error bars overlap for the open boxes meaning there is no change in selenate concentration.
In contrast pECL1e reduces selenate at a ~constant rate for the first ~35 hours and then plateaus.
They could use the protocol they used for testing the fnr gene (PCR).
Re-read the paragraph. They told you why they zeroed in on pEC223.
This is a really good figure that explains the previous paragraph very clearly.
That’s not what they said here.
They said that fnr mutants could not reduce selenate.
Propose a method from class – think about techniques you learned about in the genomics and biotech chapters.
Yes, 3 different fnr knockout mutants.
Yes. Directed knockouts like this are often used to confirm gene function. The technique is described in chapters 11 & 12.
The key here is to look up what gene the pECL32 contains (see Table 1).
What did they tell us about selenium in the intro?
Go back to the introduction and see what the authors told us about the properties of selenium and the effects different forms of selenium can have on living cells. Some organisms can use selenate as a terminal electron acceptor for growth. Are there other reasons an organism might reduce selenate to selenium? Hint: think about bioavailability.
YES!
Are there molecular biology tools you could use now that you know its gene sequence?
Not exactly.
Fnr is a regulatory protein. It turns on the E. coli selenate reductase.
E. coli has the enzyme but some strains like S17-1 lack a functional fnr gene to turn it on while other strains like K-12 have a functional regulator and reduce selenate without the need for the addition of an fnr containing plasmid.
They’re saying that fnr is absent or nonfunctional in S17-1 because adding it back on a plasmid allows “normal” expression of selenate reductase.
Look up the reduction potentials of nitrate/N2 vs. selenate/Se. That may explain the difference.
Yes! The native bacteria can carry out bioremediation (by using the diesel as a carbon and energy source) in Antarctica and they could also be useful in other locations.
Bacteria in natural environments are often nutrient limited (often N or Fe-limited). They have plenty of carbon from the oil/diesel but they lack some of the essential elements and can’t make the enzymes they need to degrade the oil. One way to speed up bioremediation in-situ is to add nutrients –essentially add fertilizer like you would in your garden.
It may have to do with the length of their field season or expedition. Many scientific expeditions to Antarctica are 3+ months long. You collect samples while you’re there and do most of the analysis when you return to your university.
Yes. Remember PCR copies a fragment of DNA. You need a forward and a reverse primer to copy (amplify both strands of a double stranded DNA molecule.
You’re on the right track. The surfactants make the PAHs more accessible to the bacteria. They act as dispersants or detergents.
In general, even bacteria isolated in cold environments can grow well (and often faster) at higher temperatures. hopefully, they’ll tell us in the next section what temperature was best for their isolates.
Happy to see no comments on this paragraph. We don’t need to know how they quantified the phenanthrene, just that they did it!
It depends on what you want to know. Sanger is preferred for sequencing a single gene (for example to sequence the 16S to identify a bacterial isolate). NGS is preferred for sequencing an entire genome but would be overkill for a single gene. We’ll talk about this more in class when we get to Genomics in chapter 7.
They’re describing the experiments that they did on a subset of their isolates here. The analyses were performed on pure cultures, not mixtures.
They’re talking about sequencing the genome here. One of the tools they used to analyse the sequence data was the KEGG database (Kyoto Encyclopedia of Genes and Genomes) which allows you to map biochemical pathways onto your isolate’s genome by sequence holology. The paper describing KEGG was published in 2000 (in the early days of genomics) but it’s been updated continuously since then.
They could test the soil and make sure it doesn’t contain diesel.
Their goal was to isolate PAH degraders so 4 contaminated sites was enough to give them a strong probability of isolating at least one degrader (we probably got >100 blue colonies from my midtown Mobile telephone pole soil!) Previous research suggests that even one contaminated sample will yield multiple PAH degrading bacteria.
Good question. Anyone have any ideas?
They answer your question in the last sentence of the paragraph – they pick those that metabolized the greatest amount of phenanthrene (i.e. the best phenanthrene degraders).
Most likely they grew them in flasks in a shaker incubator, probably at ~200rpm. This is sufficient to keep the cultures aerated. When I grew the cultures used in lab 3 I grew them at 200rpm and 35C. Without shaking the cultures would quickly become oxygen depleted and not grow as well.
Testing for biofilm formation is just another phenotypic test that gives them some more information about the bacteria. It may or may not make them better PAH degraders.
You might be able to answer why 168 hours after we see the data.
E. coli is normally a commensal of humans. Do you think it can degrade phenanthrene?
They picked colonies off a bunch of plates that they incubated at 3 different temperatures. They probably had as many or more spread plates than our entire BLY 314 lab generated in lab 2!
I’m not sure I agree with this interpretation. Have another look at the paragraph.
Two questions:
What happens during lag phase?
Are the two cultures identical? (same C source?)
They measured phenanthrene degradation by looking for a decrease in “phenanthrene fluorescence” which is fluorescence at a particular wavelength that they attribute to phenanthrene but could also come from other compounds. If the the fluorescence in their phenanthrene degradation incubations (1) stays the same or (2) increases they exclude those isolates because they (1) didn’t degrade the phenanthrene or (2) produced some other compound that fluoresces at the same wavelength as phenanthrene.
Focus on interpreting the actual data presented for now. How the bacteria acquired the ability to metabolize phenanthrene is no the focus of this figure (although it is an interesting question and on that the genomic data later on in the paper will address).
Adherence to the carbon and energy source seems like a good strategy for enhancing degradation of the carbon source. However, there are others. For example, bacteria can produce surfactants to increase the bioavailability of a PAH. They can release exoenzymes that degrade the carbon source externally to the cell and take up the metabolites. Remember the siderophores we discussed in lecture. The bacteria release them to scavenge for iron and then take them back into the cell.
Crude oil often contains heavy metals. They are not completely removed during refining.
It would be nice if they used the same units for their in-lab experiments and the soil Cd concentrations from various environments. Maybe someone will do the computation for us. You can assume that 1mL = 1g.
The beauty of tis study is that it shows bacteria capable of bioremediation are already there. They may however be nutrient limited in terms of their N, P, K, Fe etc. needs. It’s usually easier to get approval for adding nutrients (fertilizer) than for adding organisms.
During the aftermath of the Exxon Valdez oil spill one of the approached they took was fertilizing the oiled beaches. It was much more effective (and much safer for the environment) than power-washing the beaches.
Probably, remember they incubated at various temperatures during the initial isolation phase of the study.
We learn oil floats on water, right. But if enough “stuff” sticks to it it can sink once it is denser than seawater.
There’s also naturally occurring oil at the seafloor in “seep” environments where oil from below leaks out onto the seafloor. The “natural” oil spills may be a source of the oil-degrading bacteria that respond to oil spills.
The high temperatures are in the subsurface. Things cool down pretty quickly once the enter the deep sea, so sampling is quite safe.
Exactly.
Yes. When organisms consume a food (C) source some of the food is respired all the way to CO2 but some is diverted into biomolecules to build new cells.
This is something you should look up.
Surprisingly, many deep sea bacteria do fine at sea level pressure, unlike the macroorganisms from the deep sea.
Average temperature at the sea floor is 4C which is probably why they chose 4C as one of their incubation temperatures. This was also the temperature of the sediment in core 4567-24.
Based on this, any thought on why they picked 21C as their second temperature?
The oil is filtering up from below the seafloor, not down from the sea surface. remember from the introduction that the Guaymas basin contains a lot of hydrocarbon seeps where oil percolates up to the seafloor from below.
Average temperature at the sea floor is 4C which is probably why they chose 4C as one of their incubation temperatures. This was also the temperature of the sediment in core 4567-24.
Based on this, any thought on why they picked 21C as their second temperature?
Look at what they put in the flask. They put in C-14-Phe, a radio isotope, so they measured the radioactivity by liquid scintillation counting.
They ran parallel incubations, one with C-13 (for SIP) and one with C-14 to measure metabolism of the phe. Why? because they didn’t want to sacrifice the C-13 flask just to measure utilization of Phe. They wanted to save the all the C-13 flasks for DNA extraction.
They froze them after mixing them with glycerol. This allows the bacteria to be stored indefinitely. I did this with your isolates in lab 4.
Look at the temperature range for where they collected the second core.
We don’t know if they’re psychrophiles or psychrotolerant form what they’ve described so far. They’d have to test the growth rate over a wide variety of temperatures before they could determine that the bacteria are psychrophilic.
The key is not that C-14 is heavier, it’s that it is radioactive and easier to measure (and also a C-14 labeled compound is often cheaper to use in an experiment than a C-13 labeled compound as you can use way less of the radiolabeled compound as you can detect it at way lower concentrations than the C-13 compound).
You need to read this section really carefully to track what they are using the different forms of Phenanthrene for. Try drawing out a flow chart.
Basically the C-13 flasks will be used for SIP. The C-12 and C14 flasks are being used to figure out how much phenanthrene is being metabolised and if it is being metabolised to CO2 or into biomass which allows them to determine the best time to collect the C-13 labeled cells.
I’ve posted a few short videos explaining the experimental set-up in the Paper discussion module on Canvas. This diagram may also help.
Yes, the E. coli DNA will get mixed up with the unlabeled DNA but it doesn’t matter. They only want the DNA from the labeled cells (which metabolised the labeled phenanthrene). Adding the unlabeled E. coli DNA will let them determine whether they did a good job separating the labeled and unlabeled DNA.
They can test the labeled and unlabeled DNA for the presence of E. coli DNA (for example with E. coli specific 16s rRNA PCR primers).
Using 27f and 14922 reverse they’re missing abut 50/1500 nucleitides so they can’t really do any better!
What bacteria do they want to identify? The bacteria that used the phenanthrene (DNA labeled with C-13) or the bacteria that didn’t (unlabeled DNA)?
The mats are bacterial biofilms.
I’m not sure about your numbers but basically, yes. The DNA sample (mixture of heavy and light) is put on top of the CsCl solution, and then the tube is spun at a very high speed . The CsCl molecules become densely packed toward the bottom, so even layers of different densities form. The DNA moves to the CsCl layer that matches its density allowing the heavy and light DNA to separate. It looks something like the picture here.
Absolutely. Aerobic degradation is usually faster which is probably why they focused on it here.
I don’t think anything went wrong pre se. The bacteria in their samples possessed enzymes that allowed them to degrade naphthalene and phenanthrene but not the other PAHs. A general rule of thumb is that smaller PAHs are easier to degrade than larger PAHs. Water solubility (and therefore availability to the cell) is also important.
Not exactly. Your overall conclusion is correct but your pth to the conclusion is not quite right!
The DGGE gel is separating the 16S products (which are all the same size) based on their GC content. Each lane in the gel corresponds to a different layer (fraction) of the CsCl gradient used to separate the heavy DNA from the light DNA.
Look at the line graph. It shows 2 peaks, the first for the heavy DNA and the second for the light DNA.
What does the presence or absence of the E. coli band in the gel image tell us?
One would expect naphthalene (2-rings) to be degraded even faster than phenanthrene (3-rings) but biology is full of surprises!
Maybe we should try phenanthrene the next time I teach micro.
This was surprising but the difference fact that the oil exposed cores were essentially anoxic (sulfide adapted) may explain this. As Shelby said earlier, doing the incubations under anaerobic conditions might have yielded better degradation for the 4571-2 core sediments.
Yes, they are dead after the addition of the phosphoric acid.
You can google OTUs to find out! Don’t be afraid to look things up and share your findings.
11 days would be really long for our bacteria i the lab as we usually provide them a lot of carbon but for deep sea bacteria 11 days is not very long. The key here is that they chose 11 days based on the data in the graph.
You need to read the graph and the figure caption carefully. print up the graph and write on it.
The X-axis is time, the Y-axis is 14-CO2 (circles) or 14C-phe (squares).
What goes up? What goes down? What stays the same? What does that tell you?
Remember this is a paper about stable isotope probing which is using a stable isotope, C-13, to identify the bacteria using the 13C-Phenanthrene.
“Normal” carbon has a molecular weight of 12, Bacteria that grow on the 13C phenanthrene will have 13C DNA which is heavier than 12C-DNA. They can use centrifugation to separate the heavy and light DNA to identify the phenanthrene using bacteria from the
You were already exposed to this type of experiment in Genetics when you learned about Meselson and Stahl’s experiments showing the semiconservative nature of DNA replication.
They wanted to study a new site. their ship sailed to the Guaymas basin.
Hard to say if you can’t grow them! Others have managed to isolate Cycloclasticus isolates so I’m not sure I agree with that statement by the authors.
Yes, they used the acid to kill the bacteria in the sediment.
In the last paper they used E. coli as a control in their incubations with pure cultures. The only difference was the use of E. coli instead of one of their isolates.
Here, they “kill” the sediment so that the only difference is whether the cells are dead or alive. They wanted to retain the sediment in case there was something in the sediment that might react with the phenanthrene (althoug one might argur that adding the acid may have changed the chemical properties of the sediment).
That could be the case but they usually do a pretty good job mimicking things like the environmental oxygen concentration in the lab.
Yes. The phenanthrene forms a cloudy layer and s the bacteria degrade it the cloudiness disappears.
What paragraph does this comment belong to? I’ll move if and respond.
I believe I addressed this in a reply in the methods. What were the ambient conditions at their sample sites?
Check out my comments and videos on the experimental set-up. Basically the stable and radio labelled isotopes are expensive so they do some experiments with unlabeled PAH.
Think about food webs. What pattern of 14Co2 release would you expect if the 14C-PAH was going through multiple trophic levels?
It’s not important for your understanding of this paper but you can probably come up with a predicted graph.
You can buy PAH labeled with C-13. They make it in a lab.
Mostly correct! They only analyse 16S rRNA sequences in this paper. One 16s sequence dominated the sequence data. That sequence matched a sequence from a previously studies Cycloclasticus species which degrades PAHs.
They share a common ancestor in the sense that they’re all proteobacteria but they’re not very closely related. If we sequenced the genomes we’d probably see dramatically different genome content (although due to horizontal gene transfer they might also have similar PAH degradation genes).
Yes. They need a killed control so that they can be sure the changes they observe in phenanthrene levels are due to the bacteria in the sediments.
Yes. It’s very similar to “Illumina” sequencing in terms of throughput. Another NGS technolohy.
Thanks for looking this up and sharing what you found. You’re spot on. OTU is similar to a species designation.
Jordan is correct. you can figure it out by looking at the redox couple information presented in the paragraph. remember these couples are always written oxidized/reduced substance so AsV is the more oxidized substance and therefore functions as an electron acceptor, while AsIII is the more reduced substance and ca serve as an electron donor.
This would be a good thing to research and report back on but it’s not critical to our understanding of the paper.
Yes!
Not necessarily. It could just be a function of the underlying geology of Mono Lake.
They’re using nitrification/denitrification as an example or model for what happens with arsenic in Mono Lake. They are not talking about nitrification/denitrifincation in Mono Lake.
Here is a good website about toxic metals in general.
A good question that their experiments may answer.
Yes. The microbes use something other than water as e- donor. Given that they just told us about the arsenate/arsenite redox couple maybe the anoxygenic phototrophs can use the reduced member of the couple as an electron donor.
You can definitely see daily O2 fluctuations in response to light (and oxygenic photosynthesis).
A good question. Why do you think our pahE PCR failed in lab? We’re pretty sure our bacteria are PAH degraders (based on indigo production) so they should have pahE genes.
to a degree, yes. Different photosynthetic bacteria make different accessory pigments so we can use color as a proxy for the various groups. For example, in oceanography we often use pigment analysis to determine which photosynthetic groups are present.
They probably have monthly sampling trips to Mono lake as it is a well studied system. Also, one might find different microbial groups dominating at different times of year. It’s not really important for this study but there are a lot of other papers studied on this system.
You’re on the right track. Basically they’re seals that won’t pop open which is really important for their anoxic incubations..
The pH of their salt solution must have been above 9.3 before they added the HCl.
Keep it simple. Remember to focus on the big picture, not the minutiae. How they made the media isn’t really important to the big picture. The fact that they made a media that resembled the composition of the Mono Lake waters is the key detail here.
Your assumption is reasonable.
What does RFLP do? You should have learned about this in BLY 121 and BLY 302.
RFLP uses restriction enzymes to cut up DNA. You group the clones (or isolates) based on the fragmentation patterns.
Yes, they want to know if the clones or all teh same or if they represent different bacteria/archaea.
I don’t think that observation is true.
Yes, although reliable, broad spectrum arsenite oxidase primers had not yet been published).
Read more closely. They found matches for the 16S rRNA sequences. Some of the arrA and all of the aoxB clones did not yield matches. What does the latter finding tell you?
Think about how PCR works. You design primers to previously sequenced genes and use them to look for the same gene in other organisms. The arrA and aoxB primers are a concensus and not usually perfect matches. Would you expect the primers to amplify the gene from all other bacteria? probably not.
Protein coding genes are not usually as well conserved as the 16S rRNA gene so they can be more difficult to amplify.
This is a very important paragraph. The lack of comments has me worried!
Basically the pattern would just repeat again and again.
Can you explain the increases and decreases in arsenate and arsenite concentrations?
This is a pretty extreme environment (pH>9) so it’s not surprising to see low biodiversity.
Keep in mind that the bottles contain a finite amount of As that is cycling between AsV and AsIII. Also, if you look at the graph they added the As as AsIII in the beginning.
Your job is to note the changes in AsIII and AsV concentrations and figure out what biological (or abiotic) processes could be causing these changes.
Keep in mind that they’re doing this experiment in sealed bottles where substrates can run out and toxins can accumulate. It’s not a perfect simulation of the natural conditions which are more like an open container where new substrates can enter and waste can diffuse away.
Do you mean AsV reduction?
Think about why bacteria use arsenate. Would it make sense to use arsenate when oxygen is available?
Do you think the reaction is slower in the light or are we looking at a net rate? (Could oxidation and reduction occur at the same time? and is there a reason we would see the two reactions occurring in the light but not in the dark? )
Think about “who” is doing he work (need to look at the 16S and arrA sequencing results). One process was linked to essentially one species.
I think they’re simply pointing out making a connection that Mono Lake is also an extreme environment (alkaline pH rather than salinity).
Basically, they’re saying that the sulfide and arsenic are participating in many different chemical reactions and that because of this there are a variety of “As-S” intermediates present at any time which leads to errors/variability in their measurements.
Algae perform oxygenic photosynthesis using H2O as electron donor.
Yes, 50C is probably just the top limit for the Ectothiorhodospira-like bugs.
Yes.
Why do they tell us that no CH4 was detected. How would CH4 be produced in these incubations?
I think they should have stuck with their first explanation (poor primer match). It’s unlikely that there is a novel oxidation mechanism.
Think about the genomic and molecular biology techniques we learned about. Could we apply any of them to this system? Metagenomics could work as the community is not very diverse and it should be possible to get near complete genome sequences for the dominant members. Arsenite oxidase genes that did not ampplify with the primers they used might be detected by sequence homology in metagenomic sequences.
Also, arsenite oxidation was carried out by essentially a pure culture (Ectothiorhodospira-like). They take their arsenite oxidizing Ectothiorhodospira-like isolate to transposon mutagenesis and look for mutants that lose the ability to oxidize arsenite.
Loads of possible experiments to follow up with.
Changing temperatures might influence the composition of the microbial community. However, we’ll learn next week that bacteria typically have a pretty wide temperature growth range but that increasing temperature (to a point) does allow them to grow faster.
We think of the hydrocarbons as a pollutant but for some of the bacterial community they’re another food source. The bacteria that can utilize the hydrocarbons have an advantage (more food) over the ones that can’t use the hydrocarbons..
Good question. You’d usually want to process samples quickly but people often go to Antarctica for a field season (3 months or more) and don’t process their samples until they return to their home lab. Prolonged storage could definitely alter their results. Not all bacteria will survive equally well sealed in a tube at 4-8 C.
Good question. One thing that changes with depth is oxygen availability so they’d probably get more anaerobes the deeperthey sampled.
The yellow color of this metabolite is very useful – similar to our use of indigo production from indole to identify our degraders.
R2A is a popular choice in this work because it supports the growth of a lot of different bacteria. Look up the recipe if you haven’t already (I think I asked you to for one of teh pre-labs!).
No matter what media we choose we’ll bias our experiment towards some bacteria and against others. We just have to accept and acknowledge this!
moved it!
Correct. TSA is much richer than R2A which is not a good thing when you’re trying to cultivate a wide diversity of bacteria. The weeds grow too quickly on TSA!
Yes. In this case to agitate is to shake. Oxygen allows for more efficient metabolism (more ATP per glucose or phenanthrene consumed).
Yes, the M9, like our mineral salts medium, is a defined (synthetic) medium.
This is much too detailed. We don’t need to jump into how the chemical analyses work. We want to focus instead on what the authors chose to do and to measure, why they made these choices and what they learn from them.
Don’t get bogged down in technical details. Focus instead on what they’re measuring and why they’re measuring it. We don’t know enough to dive into the technical details of the chemistry methods.
This is interesting and relates to chapter 5 where we saw that bacteria can have really wide temperature growth ranges. It’s also interesting that these Antarctic bacteria grow best at temperatures they rarely see in their natural environment!
We’ll find out! SEM time is pricey so they’ll probably only look at the three highest Phn metabolizers they referenced in paragraph 3.
The last sentence from paragraph 3 suggests they’ll limit their analysis to the best Phn degraders.
Not all the isolates will have the genes necessary t form biofilms.
They’ll test each isolate separately and temperature should affect the results much. They’re already shown that most of their isolates grow at higher temperatures than are typical in the Antarctic.
There is no perfect media. R2A and M9 are typical choices for this kind of work. R2A is complex and supports the growth of a wide variety of bacteria (why we use it in lab). The M9 is defined and allows them to supply a single carbon source such as phenanthrene. The M9 also gives a “cleaner” background than R2A/R2B for doing extractions and GC-MS or HPLC analysis.
Diesel usually contains phenanthrene, which is why the authors used diesel-contaminated soils to isolate phenanthrene degrading bacteria.
Soil doesn’t have a defined chemical structure – it contains a mixture of components so we can’t really say anything about the “structure” of diesel contaminated soil. However, we can say that such soil is likely to contain phenanthrene and other PAHs.
They’re talking about sugars, not amino acids.
The key here is that the different substrate utilization patterns allow them to categorize their isolates into groups. We’ll do something similar in lab 5.
Try to avoid “why” type questions. They are good questions but we need to focus on the scope of goals of this study.
Focus on the information presented and what it tells us about the bacteria they isolated.
Remember that these isolates are members of different bacterial genera – they should behave differently. Think about the variation in height or hair or skin color we see in our classroom even though we’re all members of the same species! The bacteria in this paper should be equally diverse!
What other data did they collect about these three isolates? The answer to your question is that they probably looked at all the data and not just the phenanthrene adherence to make this decision.
The authors may address your question in another paper!
Figure 3?
What are we measuring? Is the trait/ability something you’d expect a bacterium like E. coli that usually inhabits the human GI tract to exhibit?
Note the differences in graphs A & B. They get more/better growth (higher final OD) from the diesel exposed soil enrichment.
It’s notable that all 3 isolates can grow on phenanthrene but that the diesel, which contains a mixture of hydrocarbons, does not support the growth of P. guineae (and might even kill it – would be interesting to test this).
Absolutely. Comparing the two graphs suggests that something in the diesel is killing or inhibiting the P. guineae.
Think about an oil and vinegar salad dressing. When you shake it up it forms an emulsion where the hydrophobic and hydrophilic solutions are well mixed. Bacteria often produce surfactants to allow them to emulsify hydrocarbons so that they can access them.
Their results suggest that D43Fb is not motile as it doesn’t move towards any of the substances they tested.
We might find a relative in our culture collection. Close relatives of all the isolates described in this study have been found in environments similar to our Gulf coat location.
They’re just saying that their collection of isolates is not necessarily representative of the soil microbial community in Antarctica and that an SSMS system might have favored a different subset of the community.
SSMS is a more “soil-like” system compared to putting some buffer, soil, and diesel or phenanthrene in a flask (which is what teh authors did and what we did in lab, except that we used naphthalene instead of phenanthrene).
Yes!
I like the link to what we are talking about in class! Yes, they could also take up DNA and maintain it, although in oligotrophic areas the microbes are as likely to use the DNA as a food source (lots of C and N and P in DNA).
These are way cooler – they glow in the dark. I don’t think vulnificus bioluminesces.
The key here is that iron is essential but most iron on the planet is not bioavailable – the earth contains an abundance of iron but not in a form that’s easy for organisms to take up/use (it’s typically oxidised, insoluble, etc). Paragraph 7 has more on this issue.
One thing to keep in mind is that the world was not always as it is now. Life emerged on a very different planet, one where iron was really available due to the anoxic and reducing conditions. Today the planet is oxygenated and the chemistry of iron makes it much less bioavailable under these conditions.
Siderophores are a way for bacteria to capture iron and bring it into the cell. Some bacteria make their own, others steal the siderophores of their neighbours!
The only really important thing for us to take from this paragraph is that they use two different media to grow the vibrios – a minimal medium that has limited nutrients and a rich medium that has lots of nutrients.
Everything else is unnecessary detail – important if you want to replicate the experiments but otherwise TMI.
Yes.
This is another paragraph with a lot of general information n their molecular methods. The big takeaways are that they made directed knockout mutants; they made plasmids to express some Vibrio fisheri genes ( iutA and fhuCDB).
Exactly. The plasmids they use to introduce the specific knockouts replce the target gene with an antibiotic resistance gene.
Not exactly. See my replay to Carolina’s comment above.
yes. they are randomly mutagenizing Vf to find the gene or genes that allow it to inhibit Vh growth.
“polymyxin B was added to the cultures to prevent further growth of V. fischeri ES114 while allowing V. harveyi growth. “
They tell us why they added the polymyxin. It was to stop growth (but not kill the Vf). Why would they wan t to do this? They were looking at the effect of something that Vf secreted into the growth media that inhibits Vh. They screened thousands of mutants so this was easier than filtering them which would have taken a lot of time.
Remember we looked at some of your questions at the beginning of the semester when we looked at cell fractionation in chapter 2.
Why would one do qRT-PCR?
Unlikely. Review how these reporters are made. They are on plasmids but produce fluorescence instead of the transcript (transcriptional fusion) of protein (translational fusion) when the gene or operon is induced.
Luckily they did not have to take samples every 15 minutes. the incubator/plate reader they used could be programmed to take the measurements automatically.
You could limit any nutrient and set up a similar experiment.
They didn’t set out to study iron depletion. That was just what they found was causing the inhibition of Vh by Vf. They probably went into the experiment thinking it was going to be some kind of quorum sensing phenomenon as this is a paper from Bonnie Bassler’s research group and that is what they study.
exactly.
It helps to look at the figures. In 1A +ES114 is clear = inhibited. The +MJ11 is cloudy like the no addition control = no inhibition/normal growth of Vh.
In 1b the same data are presented again but quantitatively with the black ars being no addition and the grey bars being the addition of ES114 supernatant. Some species are inhibited (Grey bar < black bar, Vh, Pa, Vc) while others (grey = black, Vp, Vv) are not.
Yes!
It means what it says – they see inhibition of Vh by the Vf supernatant when they do the experiment in minimal medium but not when they grow the bacteria in rich medium (comparison with rich media result is key). Why would the inhibition disappear in rich media?
See above and keep this question in mind for the discussion section. It is an important aspect of their results.
Yes, the inhibition they observe here is what they explore in the rest of the paper.
I would say that the different media are necessary for accuracy. The different media (rich vs nutrient limited) help define the circumstances under which inhibition occurs.
Saying that Vibrio fisheri si known to produce a siderphore… is not really accurate. to say someting “is known” implies that it was known prior to tis study. At this point all they know is that someting in the Vh supernatant inhibits Vh. They don’t know it’s a siderophore yet.
Vibrio fisheri is not using any filtration method!
The authors used filtration to determine the size of the inhibitory substance. This helped them determine whether the inhibitory substance was a protein (would not pass through a 10,000MWCO filter) or not.
The VH would eventually die but remember the staration stress response we studied in class. Bacteria can use that response and many others to survive stressful conditions by “hunkering down” and not growing, but also not dying.
This paragraph deserves some comments – it describes the first steps in how they found the genes that encode production of the “inhibitory substance.”
You don’t need to understand all the details of how they did it – just that they did random mutagenesis and looked for the loss of the ability to inhibit Vh growth.
The five arrows represent five different transposon insertion sites in the iuc operon.
Ryne, yes, iron chelation removes iron from a liquid. In this case Vf uses aerobactin to capture iron and then takes up the captured iron and uses it to grow. However, as Dayana points out, this has the effect of leaving no iron available for Vh to use for growth.
The iuc genes encode aerobactin production. We’ll see later that Vf has other genes that allow it to take up the aerobactin+Fe complex. Vh and the other bacteria that were inhibited in figure 1 must lack these genes (or have much weaker versions that are out competed by Vf aerobactin).
Yes. In your summary you should describe the data in figure 2 and interpret it in your own words. The key is in figure 2B where the iuc deletions behave the same as the no addition control with no inhibition of Vh growth meaning …
Dayana has it. The key is that the rich medium has so many nutrients that there is, in effect, no competition between Vf and Vh for iron in rich medium.
Producing aerobactin is costly so Vf only produces it when iron is scarce (minimal medium).
This paper doesn’t address whether siderophores complex other cations but they may.
This looks like the trp operon in some ways – the product of the operon (in this case intracellular iron, raher than aerobactin) acts as a co-repressor of the operon turning off it’s transcription once there is enough iron in the cell.
Exactly. These types of experiment are really important for figuring out gene regulation. The use of fluorescent reporters like mVenus, GFP etc make it much easier to detect transcription or translation than measuring the mRNA or protein directly.
Compare the activators to how AraC or CRP function in arabinose & lactose metabolism.
You’re on the right track but need to be more precise.
In figure 4A deleting luxT, yebK, fre or glpK reduces siderophore production which implies that those 4 genes play some knid of role in siderophore production – either by acting as transcriptional activators or supplying a component necessary for making the siderophore. The fifth deletion (glpF) produces the same level of siderophore as the wild type, suggesting that it is not involved in siderophore production (or there’s another gene in the genome that fulfils the same function).
Try again interpreting panels C-D (everyone, not just Makayla!)
In this case it means they did the test in strains with the exact same plasmid – one where they added arabinose to the culture to induce expression, the other with no added arabinose.
The key here is to note the genes that Vf, Vp, & Vv share and those that are found only ini Vf AND to link this pattern back to the results in figure 1 (Vp and Vv were not inhibited like Vh – now we know why).
All bacteria are able to use iron. Iron is essential for all living organisms (I think!) because of it’s ubiquity in the catalytic sites of enzymes.
The key here is how various organisms compete for iron when iron is scare enough to limit growth. Siderophores are one solution that many bacteria use. But some bacteria re cheaters and instead of making their own they steal siderophores produced by others.
Don’t forget this experiment!
The big thing to note here is that MJ11 has the same aerobactin genes (96.4% identical) as ES114 but MJ11 doesn’t inhibit Vh.
Why??? Based on what we learned about regulation of gene expression . . .
Could be due to the 3.6% aa difference is the reason.
Could be due to differences in transcription.
Could be due to differences in trabslation.
Could be due to difference sin post-translational modification of the Iuc proteins.
What do the data in figure 7 suggest?
Read through the earlier parts to find the function of the deleted genes and then think through how the loss of the genes would affect Vp and Vv. Th iuc genes make aerobactin. What do iutA and fhuCDB encode?
[By producing and secreting the siderophore aerobactin, V. fischeri ES114 improves its ability to acquire iron and, therefore, reproduce, while simultaneously denying iron to a competing species, inhibiting its growth.]
Explaining how this is done is the purpose of this paper!
This is one of the big questions in ecology. You need a balance between cheaters and non-cheaters. If everyone cheats, they’ll all starve for iron!
It’s similar to the dilemma lytic phage face where they need to infect and kill hosts to replicate but they don’t want to be too good at it or they’ll kill all the hosts and die out themselves as well.
This is also part of the equation as Vf has to devote resources to aerobactin production that the cheaters don’t but if we think about cells in a 3D environment the producers may have an advantage in taking up the aerobactin-Fe complexes just because they are at the high end of the aerobactin concentration gradient.
Agreed – it is not a simple system. It’s probably better to be a producer as you are guaranteed siderophores when you need them. if you’re a cheater you’re relying on others so a siderophore supply is not guaranteed.
This is interesting as I would have thought that one way the Vf might benefit from the symbiosis would be through supply of nutrients like iron. It’s good to be wrong now and then!
Yes, you’re correct Lataijya, it would be desirable to repress iron uptake during oxidative stress to avoid production of hydroxyl radicals.
They’re trying to puzzle out why a mutation that prevents utilization of glycerol as a carbon source would reduce aerobactin expression. They suggest that when GlpK is inactivated the bacterium has to switch to a new carbon source – amino acids. Using the amino acids for energy & growth means that they’re not available to make the protein and enzymes that make aerobactin.
This is a stretch based on their experimental data but it’s reasonable if you think about bacterial metabolism. The experiments to examine this explanation/hypothesis would merit another research paper!
Zachery is spot on.
Read on. They go through some possible explanations in the next paragraph.
They’re not saying that vents are a source of deep sea nitrate. They’re saying that as as organic matter decomposed as it sinks through the water column the ammonia that is released is converted to nitrate and that nitrate is what makes up the deep sea nitrate reservoir.
Not quite – remineralization releases ammonium from organic matter.
That ammonium can then be oxidized to nitrate by chemolithotrophs.
Bacteria that don’t require oxygen or sulfide can make a living in this zone. Previous studies have detected nitrification (ammonia -> nitrite/nitrate) and anammox (ammonia + nitrite -> N2) in this zome.
I think it’s simply a lack of studies. The sampling itself isn’t that difficult. Getting time on a research ship to go do the work is the hard part.
It’s especially important in marine ecosystems because it removes biologically available nitrogen from ecosystems that are already often nitrogen limited.
Good question. In general it is true that oxygen decrease with depth as there is no photosynthetic oxygen production to offset the consumption of oxygen during respiration of organic carbon. We’ll have to pay attention to the data and see if oxygen goes to zero in the OMZ or if there’s enough to allow aerobic ammonia oxidation.
Can you think of any process(es) that consume nitrate that could occur in the OMZ of the Black Sea?
Does anyone else find it funny/strange to learn that water layers don’t mix? These layers are not unusual in coastal systems where freshwater enters the sea.
Storms are one way that mixing of the layers occurs.
The big thing to understand from this paragraph is what “steady state flux” is, not how it was calculated.
Focus here on the data they collect rather than how they collect it. Here is a picture of the CTD-Rosette. The white object is the CTD sensor. The grey cylinders are the go-flo bottles. You lower the rosette with the bottles open and shut them at the depth you want to sample.
It may help to write out the anammox and aerobic ammonium oxidation reactions to see why labeling ammonium or nitrite with 15N can tell how the nitrogen is being metabolized.
CARD-FISH is an adaptation of FISH that amplifies the FISH signal. It’s good for natural samples with low abundance or activity which is why they use it in this study.
The CTD profiles gives us an idea if we’re lookign at a single body of water or if there are “layers” of water that might have different chemistry and biology.
This is an analytical chemistry question. We just have to trust that this is the correct way to do this analysis. The copper is being used to reduce the nitrite so that they can measure it as N2.
they have to label it as putative in GenBank because the identification is based solely on sequence homology and they have no direct functional data to say that it is an amoA gene.
What do you think generates the H2S?
yes
good
The mRNA vs amoA-cell number comparison don’t quite agree. mRNA tells one story, Cells lighting up with the amoA FISH probes tell another where the AOA are more abundant than the AOB. It will be interesting to read how the authors interpret these data.
Try writing out the equations. In (a) they fed in 15NH4. What process would yield 29N2? 30N2? How do you explain the data showing the production of both?
What consumes he oxygen? What generates hydrogen sulfide?
Would you expect a correlation between light transmission and oxygen levels?
2d is their model for fluxes of ammonium and NOx. Negative values indicate consumption while positive values indicate production. Does the model match up with the mRNA levels?
Think about the different ways N2 (4a & 4b) or NO2- (4c) could be produced from the labeled substrates they added to their incubations.
Write out the equations for nitrification, annamox and nitrate reduction.
Think about what could be driving the changes in concentration in the graphs. For example in figure 1b the decrease in O2 is probably due to aerobic respiration but could also be due to aerobic ammonium oxidation (inferred from low concentration of NH4+ and production of No3- (in Fig 1a) as O2 decreases.
1d gives us the anammox bacteria and hen ib figure 2 we can link them to ammonium oxidizing bacteria & archaea.
They are different but there may be a link between the two groups.
Compare the mRNA levels to the concentrations of NH4, NO2-, NO3- etc. in figure 1. Do you see a connection?
This is a good question. This paper is from the “early” days of measuring anammox and archaeal ammonium oxidation in marine systems. There is a lot more information now and we know that different groups dominate in different environments.
We also need to keep on mind that gens, transcripts, and enzymes don’t exist in a 1:1:1 ratio. Also, their per cell rate if nitrification might not be accurate. There are a lot of assumptions in their work.
The key thing here is that when looking only at bacterial nitrification (AOB) they couldn’t account for all of the nitrification they measured. Discovering the crenarchaeal ammonium oxidizers may account for the “missing” nitrification. (discussed in the next paragraph).
Their PCR primers are probably missing some AOA genes. Think about the PCRs we did in lab. Our “universal” 16s rRNA primers failed to amplify the highly conserved 16S rRNA gene from some of our isolates!
This is key. We often study these processes one at a time but in nature there can be intimate ties between these processes (also in WWTP!)
It would be interesting to do some lab studies at different oxygen levels to explore this more. They could need more oxygen than is available or the AOB may be better (faster) at taking up the ammonium that the AOA.
Oxygen levels are probably the key. Different enzymes may work better/worse at different oxygen levels. The data in this paper suggest that the AOA are important nitrifiers in the lower oxic zone while the gamma AOB were active in the suboxic zone supplying NO2- to the anammox bacteria.
I interpret it as the marine sequences being from the water column rather than from the sediment.
The sequences help answer whether different groups are active at different depth. It allows more insight into the system than simply measuring concentrations of various nitrogen compounds.
Right, both anammox and nitrification use the 15NH4 with the norite from nitrification used in anammox. These labeling experiments allow us to see that many different processes can go on at the same time. It’s not always an either/or situation.
It’s interesting that processes that we might have initially competed for ammonium can be coupled in this system.
I think they’re referring to places where oil is being extracted.
A lot of oil wells are located in marine systems and hydrocarbon extraction sometimes involves using saline liquids (think fracking). Therefore, understanding if halophilic and halotolerant microbes can degrade hydrocarbons is of interest. If there’s a spill, will the native microbiota be able to remediate the oil?
Microbes will eat/degrade any carbon source they can access. In marine systems they are often carbon limited and will both produce and consume many compounds we might classify as beneficial or harmful but they just classify as a carbon and energy source!
We don’t know. They’ve been found in the species listed in this paragraph but as they say “…little is known…” This paper will add to our knowledge in this understudied area.
Those of you who took BLY 314 will remember that we used MSM to isolate our naphthalene degraders. What kind of medium is MSM? What kind of medium is Luria broth? Why do you think the authors used two different media to isolate a BTEX degrading microbe? Watch the “Culturing Bacteria” video in module 1 if you cannot answer these questions.
It’s interesting that they used a soil DNA extraction kit kit instead of one for isolates given that they had the strain Seminole in culture (curious but not critical – we’ll use a kit for pure cultures in lab today).
I would guess that these were sites they already had samples from or that they had colleagues that could send them soil or DNA samples.
They do all have either “salty” or “oily” attributes (or both) so they’re similar in some way to the environment from which they isolated strain Seminole.
This experiment should be familiar to experiments we discussed in BLY 314.
If anyone took Plant Physiology you’ll remember that C-14 can also be used to measure photosynthesis.
Does everyone understand why the authors used protein concentration as a proxy for growth?
Yes. Also, there might be some bacteria that can degrade the BTEX compounds but can’t grow on them without other nutrients (for example some bacteria may lack the ability to make a particular amino acid).
Some kits work better for different bacterial species. We had a range of results in lab on Friday when we quantified your gDNA yields.
SDS-PAGE is similar to the agarose gel electrophoresis we did in lab last week. We probably won’t do it in class. We’ll focus on DNA and RNA.
Look at the figure and the figure title. The figure tells us the NaCl concentrations they used and the figure title tells us they did three replicates.
Ther could be a few reasons for the lag. #1: the initial concentration of 4-HBA could be toxic and strain Seminole may be inhibited initially; #2 – they maintained the strain on benzene but then tested growth on other substrates. Some of the substrates tested may share a common degradation pathway but others 4HBA might not and Seminole may need time (the lag phase) to synthesize the enzymes necessary to switch from growth on benzene to growth on 4-HBA.
Remember in Genetics when you studied the lac operon? You looked at a diauxic growth curve where E. coli grew on a mixture of glucose and lactose. There was a lag during the switch from glucose to lactose. We may be seeing a similar thing here.
Think back to Genetics when you studied gene regulation. Some operons (for example lac and trp) has all the functional genes near one another. Others (for example mal) has genes scattered all over the genome. It’s slightly more complicated and resource intensive to regulate a pathway that has the genes scattered all over the genome (multiple promoters rather than just one, but it probably doesn’t impose a huge cost on the cell.
Yes. Measuring both (along with the inclusion of killed controls) leaves no doubt that benzene degradation fuels growth of strain Seminole.
Those who took microbiology may remember the citrate test where we detected transport and degradation of citrate. All the bacteria we tested in lab contained genes/enzymes for citrate degradation but not all bacteria were citrate positive – Why? Because come of them couldn’t transport citrate into the cell. The could only degrade cytosolic citrate.
The lack of growth on CAT could be because strain Seminole doesn’t have the enzyme to initiate CAT degradation or it may lack a transporter to bring exogenous CAT into the cell.
Do the upper and lower pathways both start with benzene? Or does upper and lower refer to different parts of the same overall pathway?
It’s interesting that they found a gene for meta cleavage of CAT but the isolate couldn’t grow on CAT.
I liked this too. Often papers assume we know this happens but people new to the field may not know it so it’s great that our authors made it clear that the hydrocarbon breakdown products enter central metabolism.
I’m not quite sure what you mean by overlapping – maybe clarify your question in class on Wednesday??
Note: You’d usually expect all the genes in a pathway to have the same base name (all pob or all pca) but in hydrocarbon degradation there are quite a few shared pathways that were discovered in bits and pieces and at different times so the gene names aren’t always consistent.
It’s interesting that one of the regulatory proteins (PcaR) is upregulated on 4-HBA but the others (PobR, orfs 3920.3921) are not. Usually regulatory proteins are expressed constitutively.
Not exactly. They report that the paa cluster and this hybrid approach to PAA degradation has been found in many nonhalophiles. They’re simply saying that this “hybrid” strategy has now been found in a halophile as well (their strain).
It’s likely that they simply needed a higher yield protein isolation method. Also, some of the expected but undetected proteins were the smaller proteins which can be easier to “lose” in protein isolation protocols.
An angular dioxygenase would attack the 9-fluorenone pictured on the right hand side of the figure.
Note that we’re using two sphingomads in the lab at the moment, B1 and F199.
Understanding PAH/crude oil degradation pathways is certainly important here on the gulf coast. Even under normal conditions (without oil spills) we have a lot of natural hydrocarbon seeps on the seafloor in the Gulf of Mexico.
We’ll see a lot of variation in the location and arrangement of these genes depending on the organism we look at. I’m curious to see whether they are on plasmids or the chromosome in the bacteria we isolated in BLY 314.
It might be interesting to look up the sequences of the degenerate primers and see how they compare (in terms of the number of degenerate nucleotides) to the primers you designed in class.
The primers later in the paragraph are specific to the genes they found.
It may also help, once we get to the results to diagram out the sequence of experiments.
Interesting that you prefer this paper – in some ways I think the first one is easier but as you said, maybe it’s because you know more now.
Similar principles in that we’re separating biological macromolecules by size using electricity.
Agarose gel electrophoresis is much easier but sometimes you need to look for protein, especially if you make an expression clone and need to check that your host is actually making the protein.
Good to hear that it’s getting easier!
This is a good question. Phosphate buffer is almost always used for these assays. It may simply be that it was what was tried first and it worked so there was no reason to change anything. Phosphate buffer is also more similar to the media the cells were grown up in. A consistent environment is best when trying to get maximal enzyme activity.
PAH degradation genes (and virulence and antibiotic resistance genes) are often found on mobile genetic elements. We often think only of plasmids in this context but transposons are also mobile genetic elements. They can move around genomes and from plasmids into genomes and vice versa.
No. All we learn here is that the (translated) initial 267bp PCR product was most similar to a DBF63 dioxygenase. Dibenzofuran is similar in structure to fluorene so the authors decided that they had probably found a fluorene degradation gene and decided to try to fish out the full gene and maybe an operon.
Figure 2 compares the LB126 genes to those from other organisms. It’s important to note that most of these “operons” are incomplete as they come from similar cloning approaches rather than from full genome sequences.
RT-PCR analysis is one approach you could use in your projects to confirm gene function.
Will do!
Best to simply look at the table and focus on the main message – which substrates are degraded and which are not degraded. We don’t need to get into the details of the degradation products in this class – we’ll focus on the genetics rater than the biochemistry.
Protein coding genes are much more variable than genes like the 16S rRNA gene. Also, in exercise 4.2 and 4.3 you looked mostly at one genus where we’d expect sequences to be more highly conserved.
The percent yield is the amount of substrate converted to product. Fluorene was oxidized to three different products, one of which accounted for 63%, the second for 29% and the third for 7% of the fluorene.
Note that not all products add up to 100% and some add up to more than 100%. This is to be expected for a number of reasons – error in quantification, undetectable products, incopmplete transformation of the substrate, etc.
The trick is to try to simplify things. You don’t have to put all this information into your summary. Try to distill the information down to substrates where the products match CARDO and substrates where they match DFDO, and products specific to FlnA1A2. Another approach would be to score products as produced by angular dioxygenation or lateral dioxygenation.
It depends on what outcome you want. Oxygenases are used in the synthesis of some drugs because they produce very specific products – for example one stereoisomer rather than the racemic mixture produced by a purely chemical synthesis.
If you’ve ever taken Zyrtec – only the L-isomer is bioactive but I believe that the pill contains a mixture of L & R.
Enzyme “flexibility” is probably desirable for bioremediation but less desirable for pharmaceutical production.
We discussed HGT/LGT in BLY 314. Bacteria generate a lot of genetic diversity through picking up “foreign” DNA in a variety of ways. This makes up for the lack of generation of genetic diversity from sexual reproduction.
Good question. The last few sentences suggests that there are a number of possible pathways, just as we saw with fluorene.
Does Solexa yield long or short reads?
Does PATRIC generate data we can upload to IslandViewer? Does PATRIC already generate these data or something similar in the CGA?
Which do you think yields “better” data (in terms of confirming gene function) RT-PCR of an RHD gene or a targeted knockout?
Yes. There are a lot of different ways to inactivate specific genes. You probably learned about this type of mutagenesis in ~10 minutes in BLY 302! We’ll talk about various directed/targetted mutagenesis approaches in more detail in class this week.
It would be better but it can be difficult to sterile seawater and keep all the salts in solution. Also, when doing quantitative chemical analyses it is better to use a defined medium where there will be no interfering substances.
Quite possibly as in the lab we try to control everything – give the bacteria a defined medium with known concentrations of nutrients and carbon. Also, in seawater C. indicus is part of a complex community. In the lab we grow by itself.
The PATRIC CGA lessons cover this. PATRIC generates a similar map. It may help to rewatch the video in PATRIC.
The two innermost rings show the GC content (just in slightly different ways). A bacterial species typically has a fairly constant G/C content. Regions where the GC content deviates from that background level are often regions of DNA that were acquired from an external source by HGT.
The two rings with lots of different colors are showing all of the open reading frames (genes). One ring is the genes read in the clockwise direction, the other ring is the genes read in the counterclockwise direction.
Either side of the multicolored rings (and almost invisible) are two rings showing the locations of tRNA, rRNA genes and other small RNAs.
They probably used a kit like this for the carbon source utilization tests.
Perhaps. We’ll have to read on and see if they allow P73 to attack a variety of hydrocarbons or if there’s a lot of redundancy/repetition in the genome.
The arrows represent individual genes they found in the genomes. They’ve color coded the genes with the same function. The numbers below them give their position in the genome.
tRNA genes are a known hotspot for recombination in bacterial genomes. Many transposases recognise tRNA sequences.
Figure 5 partially answers Summer’s question from earlier. It maps 38 of the hydrocarbon degradation genes to biochemical pathways. One important thing to notice is that even with a complete (closed) genome sequence they did not find genes for all of the steps.
Notice too the gene numbers. These genes are found in different locations in the P73 genome.
Yes. Also worth noting that it’s best to target the earliest gene in a pathway that you can.
The level of HGT varies between bacterial species. Some bacteria are naturally competent (have DNA uptake systems in the cell envelope) and as a result often have more HGT DNA iin their genomes. However, conjugation and transduction also mediate HGT so there’s no easy way to answer your question.
Absolutely.
It looks like some of the dioxygenazses may be invovled in later steps of PAH degradation (phthalate dioxygenase and gentisate dioxygenase are probably 2 of the 6)
Much rarer than it used to be as there are now 10,000s of bacterial genome sequences in the databases. It was much easier to find a “first” 10 years ago!
Click on a bubble to post a comment about an entire paragraph.
You can also highlight specific parts of a paragraph to focus your comment or question
Think about what you’ve learned in the past about transposons. They can certainly influence gene expression (increase or decrease expression depending on where they insert in a genome).
Yes – we can use genomic analysis to identify genes acquired by HGT. Remind me to talk about this in class on Wednesday. You may have discussed this in a previous Microbiology or Genetics class. Others should feel free to chime in if they remember anything about HGT.
Alis is correct. The RHD needs oxygen. However, there are also anaerobic pathways for the breakdown of PAHs. We won’t look at them but they exist.
This is probably due to less examination of PAH biodegradation in marine environments. It’s easier to grab a terrestrial sample.
Also, many studies focus on low molecular weight PAHs as they are degraded faster and thus give results faster in the lab.
Good question. Yes, we can.
We’ll talk about this in class on Wednesday.
You are definitely missing some information, but your inferences are on target. The genome analysis identified some potential PAH degradation genes and they need to do an experiment (or three) to confirm the function of these alleged degradation genes.
Yes, they’ll need to compare their mutant to the wild (unmutated) strain to see if there are any phenotypic differences.
Reference #23 might answer this question. I’ve posted the reference list. Your question may also be answered in the results or discussion sections.
Does anyone know the coding density of the human genome?
[GI] = Genomic Island.
[Bioavailability of PAHs. Aromatic compounds, especially PAHs, are hydrophobic and insoluble in water. ]
Degradation genes aren’t enough – PAH degraders also need to be able to access the PAHs. This is true for other hydrocarbons as well.
[P73_0351 (trans–o-hydroxybenzylidenepyruvate hydratase-aldolase)]
We’ll screen your gDNA preps for this gene in the PCR lab on Wednesday.
We don’t know! Remember that in the results section they simply report their findings. They are simply telling us what they found. Authors don’t address “why” questions in the results section.
Search the accession numbers in the protein database at GenBank here to learn about the different enzymes they list here. We know these numbers are for protein sequences because the names start with a capital letter and are not capitalized. Gene names start with a lower- case letter and are italicized.
The Nitrogen replaces the normal atmosphere, making the bottles anaerobic.
We’ll have to see the results of their Blast analysis to see if your assumption is valid. The sequence matches may indicate if the sequences came from bacteria or archaea or both (or they may not be able to determine this). Can you think of a way to determine whether Mono Lake bacteria, archaea, or both contain arrA?
We analyzed this figure in class on Tuesday. Watch the zoom recording if you’re having trouble interpreting the graph. Figures like this make good exam questions.
We analyzed this figure in class on Tuesday. Watch the zoom recording if you’re having trouble interpreting the graph. Figures like this make good exam questions.
The paragraph title tells you why they added hydrogen. We discussed this in class yesterday. Watch the zoom if you have trouble with this paragraph. Basically they are trying to determine what electron donors the Mono Lake microbes can use (and what electron acceptors).
Think about what is serving as carbon source, electron donor, energy source and terminal electron acceptor in the various incubations.
They identified the arsenite oxidase gene from the arsenite utilizing phototroph a few years after this study. At the time this work was done there were only about eight published arsenite oxidase gene sequences which made designing primers that would target a broad spectrum of arsenite oxidase genes challenging.
That’s exactly what happened. Any time you use degenerate primers for a protein coding gene there’s a danger they will amplify the wrong target and give you incorrect/nonspecific results.
Keep in mind that the mat was dominated by Ectothiorhodospira, so it’s not a total surprise that the diversity of functional genes would be limited.
I was interested in the use of these microbes in cleaning up mass pollutants in the environment, and the process by which it is done was interesting. Essentially, workers will attempt to create an ideal environment for PAH degrading microbes to flourish in by adding in carbon, nitrogen, and trace amounts of phosphorous. They will also attempt to increase the surface area of the oil using biosurfactants so that the bacteria might “attack” the particles easier.
Well the phosphorus would make sense as it is a key part in cellular respiration, this would stimulate the activity of any microbes. Nitrogen, however, seems like it would be useful towards any phytoplankton that might break down PAH. So it would make sense that these inorganic molecules would enhance PAH breakdown since they are encouraging the growth of the microbes. At least, this is my understanding of it.
I’m not sure about the time needed, but I do know that 180 RPM at room temp ensures that the bacteria grows an disperses throughout the sample evenly. Perhaps the two weeks is simply provides the best results? And we practice much shorter time frames for convenience sake.
Without reading the paper or reading anyone else’s comments, I think I can derive the meaning of the data displayed in the graph. The control group appears to be a medium with no microbes present. The higher rate of Fla degradation in the YMSM plate tells me that the YE raises the degradation potential of R. erythropolis.
Ah, now this seems vaguely familiar…
My question is in regards to homology. This paragraph states that the colony exhibited a 99% homology with Rhodococcus. Is this from random mutations that might occur in growth? And what is considered the cutoff the colony to no longer be homologous?
This figure is rather confusing to me. Having the 100 and 250 concentrations totally degraded and then nearly 50% of the 500 concentration makes sense following the trend. But then the 1000 mg/l only saw 12.6% degraded. I wonder what could cause this? Does an abundance at that level mean that it is lethal to the bacteria? Is there so much in the environment that there is no need to compete over it?
I think its interesting that it not only can be used as an herbicide, but I did some researching and found that it can also be used to degrade herbicides! Im looking to see if it can also be used to degrade pesticides, and if so it can create serious benefits for our environment.
I’m happy to find that Rhodococcus can be used to degrade PAH’s! With oil spills, overuse of pesticides, and general pollution happening every day, it helps to relieve me knowing that we are finding new ways to deal with the constant pollution we are causing.
Disregard my previous comment, it was for the last paragraph.
I’m still curious as to why an increase in Fla concentration resulted in an inverse rate of degradation. I would assume that it is either a lethal amount of Fla, or perhaps that there is so much that the microbes no longer need to “compete?” Though I’m not sure how much scientific basis that last thought has.
Well, at least I Don’t eat seafood!
Still, the process of using microbes to clean up pollutants in an environment seems to be a lot more prevalent than I had initially thought. When I have more free time, I think I’ll look a bit more into the practice and see what other types of uses this can serve!
Perhaps the concentration of carbon in the water might have an impact on its ability to break down these carbon groups? I’d imagine that C is much more available in the air than the sea, and this would explain it if they were more eager to break down any source they could find.
A defined medium would be used in this instance because the researchers were trying to isolate a DNA sequence for a specific strain. Therefor, it would be better to use a medium where all of the ingredients were known to promote the growth of what you want and inhibit any unwanted strain. If this is correct, does that also make this a selective medium?
I’ve had to dust off all of my old chemistry notebooks just for this class! To refresh on the differences between ester and ether for the cell membranes, re-learning redox reactions, looking up what analytical techniques entail, its a good thing I saved these.
Adding onto this, why is the exact same amount use for MeOH as the medium? I can’t find anything that would suggest an answer.
I’ve always felt that evolutionary biology was my strong suit, so I am no stranger to phylogenetic trees. That being said, I’ve only worked with larger scale organisms and never anything microscopic. It looks like the basic flow is the same, but genetics play a much bigger role in diversifying microorganisms since phenotypic observations can only go so far.
I believe this should have gone under Paragraph 8, apologies for the mix up.
No, THIS should go under paragraph eight.
Does a genes location on the strand carry any significance? I tried looking around but all I can find is discussing gene locations in regards to knowing where to look rather than function. So it seems like the locations isn’t as important. But still, it is interesting to see the location vary!
An ISP is an iron-sulfur protein. And aside from forming the structure of the aromatic deoxygenase, I’m also having a hard time understanding tis specific function, if anyone has any input I would appreciate it.
ISP is iron-sulfer protein. And aside from serving as part of the structure of the aromatic deoxygenase, Im having a hard time understanding tis specific function. If anyone has any input I’d appreciate it!
While I knew that the microbes that live in thermal vents like the ones in Yellowstone and Paoha Island were extremophiles that thrived in hot conditions, I never thought about their nutrient sources (or much of them in hindsight). I never knew these thermal pools were toxic too, and I’m interested in learning more about the metabolic processes of these bacteria as we read the paper!
Well if we assume an extremophile is simply an organism that can survive in conditions that are unfavorable to humans, I would say yes based on the fact that even if PAH’s cause harmful effects after repeated and longer exposures, they still are an environment humans cannot live in long term. So I think it would have basis to say that PAH degrading microbes count as extremophiles.
Well the color of the hotsprings would suggest a different chemical or microbial makeup compared to the others. zAs for why detail was went into for the location and other geological features, I would imagine that all kinds of data is recorded in regards to the location of extremophiles and where they are found. Certain features might hint at certain chemical makeups of the environment, and it never hurts to be precise and meticulous in gathering details!
Did they combine the products so that they could submit the final product to BLAST and compare the clone sequences to one another? Is that something that can be done?
It makes perfect sense that we would see more archaea than bacteria since this is an extreme environment in regards to chemical makeup and heat. In addition to this, I feel like this is one of the easiest papers to read so far! Im not sure if its because we did all of this firsthand in lab or its the third time I’ve read one, though. Maybe a bit of both?
Im having a hard time understanding why the amount of light has any affect on the reduction oo oxidation? Are these bacteria photosynthetic in nature? Does the amount of light have an effect on the electron exchange?
It makes sense to me that MSM plates are commonly used when growing cultures of bacteria. After a bit of digging around, it seems that this is because salt essentially drains all the water out of the microbe if it is overexposed. The textbook tells that a a bacterial cell is majorly made up of water, and the MSM plates used in lab 2 grew too many colonies to count. All of this makes sense, and it seems the minimal salt plates are favorable to grow large amounts of microbes, but not singular colonies.
Also I have no idea why the profile picture decided to pull from my old social media page…
What is the normal ecological niche of these bacteria and why would they specificly have a specialization to degrade PAHs? How universal is the gene to produce these enzymes? Is the gene on a plasmid and easily exchanged through conjugation or are is it part of the chromosomal DNA?
Where did you read that PAHs expedite growth and increase population rate? I suppose that if we are able to isolate these bacteria using napthalene enrichment media that they are able to use the PAH as a metabolic energy source but are they actually growing more efficiently then if they were using another energy source? Perhaps it takes a longer span of time to completely eradicate a polution problem in the real world due to environmental factors (access to the pollutant, other carbon sources for food, pH, etc) that would be controlled for in the lab?
I understand that the BATH assay tests how hydrophobic the bacteria are but what does this tell us about the bacteria in relation to being naphthalene degraders? Is this just a standard diagnostic test or does it have special relevance in this case?
I wouldn’t think this was necessarily to “characterize” the naphthalene degraders but nowadays you can’t really describe anything without molecular analysis, right? And knowing what you are dealing with in relation to other genomes helps us give a name to what we a working with or possibly even describe a new species.
All the specimens showed sufficient growth compared to the (positive?) control which was simply stated as a physicochemical degradation but they did not describe what process exactly they used as the control.
The cloudy white substance is actually the emulsion of two liquids that usually do not mix. It is what they are measuring in the emulsification test. An emulsion may look kind of soap bubbly to opaque white like mayonnaise. They measure the size of the emulsion compared to the total volume height to get the emulsification activity ratio.
I believe the more effective degraders (even if they have lower growth rate) are better emulsifiers and have higher hydrophobicity. But just because you can degrade it faster doesn’t mean you also are the most efficient at turning around the carbon into biomass. That being said, I think it would be interesting to compare the size of these particular species and see if volume to surface area does play a role.
Has a study been done to see if asphalt and resin do inhibit gram positive growth or was this an assumption on their part? Obviously some gram positives are capable of naphthalene degrading but to make the statement that gram negatives “better tolerate and uptake PAH” than gram pos due to cell wall structure shouldn’t a more specific comparative expirement be done? (I read the Tebyanian paper cited here and saw no refernces to cell wall differences and in fact the conclusion to this paper was that their gram positive Tsukamurella was the better degrader of hexadecane which is not even a PAH)
Perhaps the gram positive’s thick peptidoglycan membrane (instead of the gram negative’s two membranes) allows it to be more adaptable to a constantly changing environment?
This research, and most of the cited papers, deal with marine bacteria. Do the traits of being bioemulsifier/biosurfactant producers and optimal growth at 400 ppm apply to soil bacteria as well?
This is the first time I’ve seen substituted PAHs mentioned. Are substituted PAHs more difficult to degrade/use due to the constituents of the side chains or is it relative the size of the overall molecule?
Are we expecting the enzymes–and genetic regulation–used for the degradation of these PAHs found in a marine species to be significantly different from those found in terrestrial bacterial species?
They used indole crystals as an indicator to differentiate the E. coli that contained the Cycloclasticus genes for naphthalene degradation, just like we do in lab. E. coli are not normally PAH degraders (I assume) and so they can be confident those expressing the indigo are carrying the genes of interest.
The E. coli is grown on agar supplemented with antibiotics to select for only the bacteria with recombinant genes. This is possible because the researchers included an antibiotic resistance selection marker in the formation of their plasmid. Standard practice in this type of experiment but was not explained.
AUG is the RNA analog of ATG. Remeber that uracil is used in the place of thymine in RNA.
Overall the percent identity matches are low, coupled with the divergence from the nah-nod-dox-pah genes mentioned later, suggests the Cycloclasticus genes are not closely related to other known genes for pah degradation.
The way the previous paragraph was written is a little misleading. They did have a gene for the small subunit, it just was not located in the “normal” flanking region paired with the large subunit gene. If you look at table 3 and the dendrogram again you will see phnA2 was what they were designating the small subunit gene.
How would we test whether the proteins are expressed constitutively or coordinately? By growing the strain on substrates with and without PAH and then performing an enzyme/activity assay?
So, if I am reading it correctly, the PhnA2 is the beta-subunit partner of PhnA1. Together they make the functional ISP (iron sulfer protein). PhnA1b is only an alpha subunit and is missing its beta subunit and thus should not be functional based on previous research. If it can bind to this other protein’s beta subunit then it could still be useful to the organism. Otherwise, is it just a leftover bit of gene that doesn’t do anything?
These bacteria use As (III) as the electron donor instead of water and so the waste product is As (V) instead of oxygen. They still perform photosynthesis as they still convert light energy into chemical energy, there are just different chemical reactants and products.
I also read that anoxygenic photosynthesizers use a different type of chlorophyll than oxygenic photosynthesizers.
Are the Archaea found in these biofilms part of the As (III)/As (V) biochemical cycle or are they serving some other function in the biofilm community?
What was the purpose of bubbling with nitrogen gas before sealing the samples? Can these bactria fix nitrogen?
If all of the aoxB clones failed to show similarity to anything in Genbank, does this suggest a problem with the primers used or might it have been a novel gene sequence not previously described/uploaded to GenBank?
When reduction of arsenate is driven by H2 or sulfide, with their higher reduction potential, more energy is released that can drive carbon assimilation into biomass. In the light incubation the biofilm seems to be busy oxidizing arsenite which doesn’t give off energy to drive assimilation. Maybe another process was assimilating the carbon that didn’t have to do with the arsenic oxyanions?
Why does the oxidation of arsenite appear to occur faster in the dark incubation sample? Is the chemolithotrophic metabolic pathway quicker?
What is the rate at which bacteria grow due to degradation of PAHs? Is it a significant amount of growth to the bacterial population?
Since PAHs expedite growth of bacteria, thus increasing the population rate, why is there such an excess of these pollutants? Is it due to the fact that the enzymes present, even in larger populations of bacteria, are supersaturated with the pollutants – meaning too much to degrade entirely? Or are there instances where the bacteria is enough to eradicate the problem?
Why were these specific wavelengths used?
Is 94 degrees Centigrade not high enough to denature the DNA, protein and enzymes of the bacteria? What is the optimal temperature range for the bacteria?
Why do strains N1 and N7 exhibit such high levels of degradation when according to their phylogeny they are of very distant relation?
Can a reliable trend and correlation be determined from the data? It is leaning toward a trend of positive correlation, as the E24% increases, the BATH% increases as well. However, there are many that do not follow this pattern linearly.
why are two-ringed aromatic compounds easier to decompose? Is it due to more steric hinderance in the electron shells making the bonds easier to break?
I think it would be the cell wall because it has that extra layer of protection/barrier causing the cell to handle different environments.
In general, how much do the bacterial populations from marine environments vary? The previous paragraph discussed sediments from Tunisia, how different are they from the populations else where, i.e. are specific/unique species in certain locations?
As stated in the article there are 54 different bacteria, what are the specific advantages for these bacteria? Are they not all competing for the same nutrients or is there so much contamination that allows this level of diversity with out worrying about competition?
If seafood contaminated with PAHs were treated with PAH degrading bacteria and sterilized, will harm still be inflicted on human consumption of the newly sterilized seafood?
How different are the genes in degrading different naphthalene molecules?
I had a similar question, except that, were they used interchangeably?/ or was an entire experiment given with maphthalene or phenanthrene?
why bacteriophages? Is there less variablity and more guaranteed that transduction will occur? Is transformation and conjugation less likely to occur or not as easy to measure if it were to occur?
Is this gene required for all naphthalene degrading bacteria? Are there naphthalene degrading bactiera that do not have the ability to oxidize indole to indigo?
At what percentage is considered too low to be considered a different function of the gene? How much can be based on variation and genetic diversification versus random coincidence in amino acids?
Why does it appear that plasmids are more likely a source for PAH degradation genes as opposed to the chromosomes, when it comes to the species found in the environment? Is that specific for this enivornment or PAH degradation as a whole?
Is it only a coincidence that the beta subunit gave a tree similar to that constructed for the alpha?
Does the spacing of genes in relation to one another play a significant role in function?
Is it uncharacteristic for a species to has more than two kinds of a dioxygenase genes?
is it specific for bacteria that utilize arsenic to form a biofilm? or is that only specific for this particular bacteria?
Is Arsenic utilization dependent on the presence of oxygen?
Is there a reason red algae had lower temperatures?
why was HPLC used? IS this the only way to measure this variable?
Are these Archael strains also anoxygenic photosynthetic oxidation of arsenite?
Could it be possible the the energy accumulated in the photosystems from the photons of light take a while to de-energize during the dark cycle, hence why it is a longer period?
why ws a radioisotpe not used the entire time in the experiment?
For a practical sense of research, how useful is this. Due to the fact that different environments will have different bacterial systems and biofilm formations?
When it states exploring what electron donors, is the environment exposed to different compounds or will these temperatures or time changes cause a pronounced change in the electron donors itself?
Do bacteria no longer have these light and dark rhythms?
It is amazing what you can learn from just reading this first paragraph so far. I knew, from what I have learned in organic chemistry, that cyclic aromatic compounds could either be harmful or good but I never know that they affected us this bad. I never really thought about it on such a small scale that goes all the way done to the make of different pollutants.
If the organisms needed to resolve matters like oil spills and other chemical pollutants, why does it take so long to resolve the matter? Also have there been changes to procedures to help ensure that problems like this do not happen or are taken care of in a timely manner?
What makes ONR7a medium different from R2A agar? Why was ONR7a used instead of R2A?
Will we ever use the Big Dye terminator V3.1 cycle sequencing kit in lab? How often is it used in the field of microbiology?
I would like to know what emulsification activity is. I did not quite understand what it was or the purpose it served when I read the materials and methods section.
Will we be utilizing the National Center for Biotechnology in lab as the semester progresses?
Are there naphthalene-degrading bacteria in the gulf coast near the oil rigs that produce biosurfactant and bioemulsifier? Are there more now in the gulf coast compared to before the big oil spill?
Where would we find more Gram positive bacteria versus Gram negative bacteria?
Why does 2,3-dioxygenase mirror “the taxonomic grouping of the host bacteria”?
Are naphthalene-degrading bacteria used more than phenanthrene-degrading bacterial in most labs? Would phenanthrene-degrading bacteria have an effect on culture samples in lab, depending on if there are certain oils present?
What is Sall? I was able to read up on Takara and the work that is done through Takara. I could not find anything related to Sall.
Does Sall make it easier for the primers to attach onto DNA? Is DNA manipulation related to PCR?
In microbiology, would it be more efficient to use DNAMAN 4.0 software or MEGA software when it comes to phylogenic analysis?
After looking in the genus Sphingomonas I found that thy are all gram-negative and rod-shaped. They are either yellow or off-white in color and have about 37 species.
Why can’t phthalic acid and diphenylamine support the growth of ZX4? Is there a specific compound in them that prevents growth?
Are there specific bacteria in the genus Ectothiorhodospira that they are looking for or they just interested in the fact that they are purple sulfur bacteria?
Are there other samples they could have been taken from a different area that would have worked with this experiment and given similar results?
What was the reasoning behind the dark-incubated controls and samples from arsenate reduction experiments being wrapped in aluminum foil?
Are radioassay incubation and radioimmunoassay similar? I could not find anything about radioassay.
Arsenate respiratory reductase is also found in Desulfosporosinus strain Y5. They are spore forming bacterium.
While doing some research on Ectothiorhodospiraceae I found that they belong to an order called Chromatiales. In that order there is a genus called Halothiobacillus that is not photosynthetic.
This paragraph does a really good job at setting up the current issues and importance of Antartica to our global climate. Selfishly, I would love to go and explore the wildlife that I’m only able to see on National Geographic, however, if the cost is this high should tourism even be allowed? There must be some sort of regulation in the future until we are able to leave less of a footprint while visiting as tourists.
It is actually really amazing to me that a place such as Antartica would be so well equipped with NAH degrading bacteria species since their would typically not be a huge use for them. That being said it seems like it is a race against time because the scientists themselves must be using some sort of fuel, most likely diesel, so a really strong NAH degrading bacteria must be found before it is too late. Is biostimulation already in use?
While reading this the question came to mind, does this area have a deep layer of permafrost (I assume it does)? If so are the PAH degrading bacteria located in it? Or is there an active layer of it that the bacteria are located in?
I wonder how they quantified areas as “diesel fuel-exposed” or not. Did they pick soil areas that were in close proximity to ports or roads? Also, I was wondering why they only chose three strains from the 53 that were PAH metabolizing?
I was able to understand the explanation on the results but got really confused when looking at the figures. I am mostly confused as to what is going on in the graphs in part G and H. I can see how D43FB has the highest degradation in figure A but is then outdone by PAO1 which I think is just something to compare it to in the lab, and probably a more common and well-researched bacteria.
It seems unprecedented that a bacteria could be so well adapted to not only be a great degrader but to resist heavy metals. This must have been extremely encouraging for the scientists. I wonder if prior to the experiment they had any idea that they would find this bacteria in the soil? Had it been discovered or researched before this project?
I was wondering if this work was followed up by any bioremediation techniques? Is there not enough information in this research and previous research to support bioremediation using D43FB? I know that they are all aware of the current issues and the urgent need for bioremediation but do more tests need to be done before they are allowed to use bioremediation in Antarctic regions?
The more I read about the soil collected, the more I am amazed at the diversity of the soil in this region of the world. When I think of Antarctica the last thing I think about are what types of bacteria are in the soil. It is very interesting to wonder how some types of bacteria are found very far away from each other in similar conditions.
I’m wondering if bioremediation of the sea floor could work the same way as bioremediation in the arctic. I am also curious about if there were any bioremediation efforts immediately after the Deepwater Horizon spill.
I’m wondering how the scientists were first able to find these various deep sea environments. Also how are they able to collect samples from such places that are over a mile deep in the water and are such harsh environments (pressure, temp, etc).
I was wondering about the change in temperature. Since it said the soil at lower depth was found up to almost 50 degrees, storing it all at +4 C, wouldn’t that interfere with the microorganisms growing at depth. Or are they irrelevant to the study?
Just out of curiosity, I was wondering how long it might have taken these oil droplets to get into a soil sample like the one that is pictured? It seems like it would take a very long time to not only filter down through over a mile of water and then into the soil.
I’m also wondering about the ability to degrade PHE just like in the arctic. There it was due to increased human activity. Is this area of the ocean somewhere where there is frequent traffic of ships using diesel fuel? Or potentially near an area where there is a lot of offshore drilling rigs?
I was reading and looking at Fig3 but I’m having trouble making sense of what the circles are actually showing being measured.
Since some bacterial taxa have been identified that have the ability to degrade PAHs will there be similar bioremediation efforts such as the efforts used in the arctic regions? I was also wondering if deep-sea sediments were the only place where such bacteria are found. It seems like heavily commercialized areas such as marinas could benefit from the same bacteria in much more accessible and shallow waters.
In our last paper it was interesting to me that the same species of bacteria were found on different sides of the globe even though they were found in similar conditions. For them to find the same bacteria in different regions of the sea floor makes more sense to me because the ocean is all interconnected.
I was wondering why these oxyanions are found in anoxic biofilms? There must be some correlation with the oxygen level and presence of the oxyanions.
It is interesting that they found so many different metabolic activities being used, especially with a compound that is usually thought to be deadly. It goes to show how interesting microorganisms can be.
I was wondering what gives the red coloring to the biofilms in the springs? I have never seen hot springs but when I think of biofilms involving water I think of the green biofilms that are characteristic of a lot of ponds and swamps.
Morgan, I was also wondering why the clones did not show up in GenBank. Maybe they mutated or the isolation process was not exactly right and gave them a different result than what they were hoping.
From reading this I thought the same thing that you did. It seems like natural conditions would favor the reaction occurring for there to typically be a higher concentration of As(V).
Since reduction and oxidations happen in coupled reactions, this graph seems to illustrate that point. I do agree with you though it seems like the reactions take longer in the presence of light.
In some ways i think it could be disappointing that they found no results but also exciting that they might have discovered something new
I wonder where else on Earth similar ecosystems could be found, and if they harbor the same microbes or different ones.
Like Julie, I looked up more specifically where PAHs come from. For factories that burn coal or where exposure to workers are concerned, or even cities where burning these materials are present, I wonder what the percentage of disease is compared to a population that has minimal exposure. Based on this first paragraph, it would be indicative that disease percentage would be higher in an exposed population versus an unexposed or minimally exposed population. That being said and with research available, I wonder what safety precautions or measures are taken to protect factory workers. Would a certain type of mask suffice? Would burning something like coal introduce PAHs into the air that would then settle on maybe a nearby field that has a food source, which can then be ingested by animals or by us?
Maybe I missed something, but would a LMW or HMW for PAH-degrading microorganisms mean something, as far as use? Would mentioning it pertain to economic cost for production or transport?
I wonder if these enzymes could be isolated or used as a model in some way and then used to degrade PAH. However, if there are no adverse implications to using Rhodococcus, I suppose using them to degrade PAH would be self sustaining and more economical. I would be curious to see how else they would interact with their environment.
Here a 5% NP and 1% of the other PAHs were used, but I wonder what a higher percentage of those sources would do for the experiment? Would it be beneficial or pointless? Also why were alcohols used?
Was the MSM plate used at first to encourage a general type of bacteria to form? In the book, it says that an enrichment culture is used to select for certain microorganisms. Certain media and incubation conditions are also used to select. I suppose the colonies transferred to LB would be selected for. Also, it says that the colony CMGCZ was inolculated in LB and incubated at 30 degrees Celsius and 180 rpm for 48 h. Why was it spun (180 rpm)?
The Fla degraded quickly in YMSM, but I wonder why the MSM took so long? And why did it quicken after the fifth day? Why would removing Fla change this?
Does this mean that if CMGCZ was commercially used one day that it would not be able to handle large amounts of Fla or would CMGCZ have to be added in more abundance? Perhaps the goal isn’t to just use CMGCZ but rather just use it as a model that can later be developed to handle more Fla.
I’m interested to know how Rhodococcus or even CMGCZ would interact with other things or organisms in the environment. Would the Rhodococcus be released to do the job of degrading PAHs and then be done or would it have any negative effects on the environment? Perhaps the perk of PAH degrading would outweigh the consequences, whatever they might be, of not having a degrader.
I was researching PAHs a little and found that LMW PAHs are acutely toxic, while the HMW PAHs are mutagenic and carcinogenic. This research seems promising in that LMW PAH degraders are more successful in research, since LMW PAHs are an immediate concern, especially to aquatic organisms.
I found that PAHs are particularly a problem when mixed in with soil where they build up and are not easily obtained once there; aquatic environments seem even worse off. Here we have the mention of oils. If the oil were to spill in the soil, or even in an aquatic environment, like in the BP oil spill, how would this increased difficulty of PAH breakdown be addressed? Perhaps future experiments would show the promising PAH degraders in environments such as these.
In a couple other studies I’ve look at, fungi and bacteria like Rhodococcus were used to degrade PAHs, and while the bacteria did well in degrading LMW PAHs, the HMW PAHs proved to be the problem. The bacteria seem to only partially oxidize larger rings. With the fungi, PAH degradation is not their ideal source of carbon, but they will degrade the PAHs to a detoxified metabolite. Perhaps a mixture of both bacteria and fungi could help solve the problem? That is, if they’re compatible and not harmful in combination for the environment.
It is interesting that grass fires produce PAHs. If I recall correctly, our local area with Weeks Bay Reserve performs clearing of certain areas using fire. It would be interesting to observe the amounts of PAHs in the nearby waterways, especially the dead end canal areas. Perhaps they take this into account and use something to degrade PAHs?
I was looking into how PAHs were produced, and we know that now they’re mostly produced by industrial activities, but I was interested in how these bacteria came about to degrade PAHs before industrialism or synthetics came into place. Perhaps there was an organic source? As it turns out, PAHs can be produced biogenically by plants and pyrogenically, like the article suggests earlier. Biogenic PAHs are produced by the breakdown of vegetative matter and get trapped in sediments. Interestingly, biogenic PAHs tend to have more rings compared to petrogenic (from industry) rings. Different sources of PAHs also have different ratios to what kind (ex phenanthrene vs anthracene ratio). It was a little unclear as to why the breakdown of vegetative matter would produce PAHs, which would later be utilized by PAH microorganisms. However, the different sources of PAHs and different structures produced by such were interesting to read about.
I wonder what using a mixed, but filtered sample of PAHs from an environment would do? Use the purified ones here, and evaluate results but then use a mixed one from the environment as a step forward from the isolated ones.
It seems that 1-methylnaphthalene has a high conversion rate. What does the methyl substituent contribute towards conversion that the plain Naphthalene lacks? Also, why does placement matter, in comparison to the 2-methylnaphthalene?
pH1a and pH1b had the ability to oxidize indole to indigo. Does this mean that our degraders might have these constructs as well? Or perhaps, how common are these specific constructs in degraders able to oxidize indole to indigo?
I wonder if the phnA4 gene, which has an N terminal region similar to the chloroplast ferrodoxins, are somehow related or came from a common ancestor? Also, it is interesting that P. stutzeri was mentioned here. When I looked it up, it said that it can be found in human spinal fluid, and here, it is mentioned that phnA4 has 47-51% similarity to the NADH-ferredoxin oxidoreductase components. That is interesting that they are so similar and I wonder what the history of this similarity is.
We have plasmid versus no plasmid. I wonder if the Cycloclasticus sp. strain A5, somewhere in it’s history, had a plasmid or fragment of DNA copied into its chromosome, like an episome?
I suppose the difference in location, sequences, and clusters would indicate that different regulations between different microorganisms came into play, even though the basic features of the protein family were conserved.
It is interesting that in E. coli coexpressed alpha and beta subunits were unproductive, but in Cycloclasticus sp. strain A5, the opposite was true. It seems here that the close, “contiguous” placement of the alpha and beta subunits with the genes needed for electron carrying are placed rather specifically. This seems efficient and concise.
Here it mentions that the three dimensional structure of the NP dioxygenase of Pseudomonas sp. strain NCIB 9816-4 has a long narrow gorge which allows it to be able to break down the PAHs that it does. This is an interesting mental picture of what is going on with some of these bacteria. It then mentions conserved residues and divergent ones in the channels in Cycloclasticus sp. strain A5. The diversity of Cycloclasticus sp. strain A5 may contribute to its ability to break down various PAHs. Earlier, it mentioned that Cycloclasticus sp. strain A5 did not have a plasmid, but I wonder if this genetic information coding for this ability would be able to be excised (Ex CRISPR) and placed into a plasmid (like from some of the other bacteria mentioned here) in order to try and get other bacteria to take it up? Or at this point, we wouldn’t even need a plasmid; rather just CRISPR and specific placement of genes.
If primers were the issue, perhaps they could study other arsenic using organisms, use transcriptomics or proteomics to help identify genes and usage of such; and by doing all of this, perhaps they could come up with another primer that would be a success? Perhaps that is too naive to think, though.
I did a little research, and like the PAH degrader investigations, researching arsenic utilization would also be useful to us in breaking down toxins. Orchards, due to fertilizers and commercial treatments, tend to have a lot of arsenic, as well as other inorganic compounds present in the soil. Knowing this, it would be interesting to not only study arsenic degredation but how that metabolism is affected by the presence of other inorganic compounds.
Did they choose those specific times for sampling because of environmental changes (hot environment, cold environment)? Even so, the water is noted to stay pretty constant, near 45 degrees Celsius. Would a more consistent study use a more consistent spacing in time, or even sample once, every month, during the same time of day?
This is an interesting detail. Perhaps the toothbrush was used just to separate the microbes, while the spatula was used to keep the DNA intact. I am not sure, though!
Perhaps there is a difference of initial rate of change from oxidation and reduction states in the light vs dark reactions because, like we discussed in class, there was a selection for a characteristic that normally would have been selected against (fitness wise) in a light environment; but it became advantageous for fitness in the dark. I would be curious to see how many generations passed, the rate of change for the population, and a proteomics or transcriptomic study to compare the differences between the microbes in the light and dark.
I would think that they could use a primer from a known organism that does have the aoxB-independent mechanism and use that as a primer for the aerobic As (III) oxidation activity. I am surprised that they did not do this, but perhaps that just gives them a reason to do another study.
I may have missed this but does the light driven As(III) oxidation have such a narrow range for temperature at 50 degrees Celsius because that is maybe what the temperature would be in its natural environment?
Perhaps the temperature range is so wide in the dark because the presence of physiologically different anaerobes are the more important factor here. Maybe this allowed the microbial population that does light driven As(III) oxidation and dark As(v) reduction to produce energy day and night without interruption.
I think they just wanted to see what electron donors best fit their studied organism and avoid bias based on what they suspected. It’s probably just for clarification an confirmation.
Were they expecting to find that chemoheterotrophy drove As(V) reduction? Do these results seem to suggest that chemolithotrophy or photolithotrophy might be more responsible?
PAHs are found in meats cooked directly over a heated surface. Juices from the meat drip downward into the fire creating PAH filled smoke that then adheres to the meat.
https://www.cancer.gov/about-cancer/causes-prevention/risk/diet/cooked-meats-fact-sheet#what-evidence-is-there-that-hcas-and-pahs-in-cooked-meats-may-increase-cancer-risk
With PAHs having harmful effects on the respiratory system, I wonder if there’s a difference in the amount of exposure to PAHs between cigarettes and vapes not only for those who smoke, but also for those who inhale the smoke secondhand.
Is aoxB the only known gene that codes for aerobic As(III) oxidation?
I find it surprising that the biogeochemical cycle of arsenic is not as studied what with arsenic being found in high levels of groundwater and it being highly toxic and carcinogenic.
So microbial degradation will remove the toxic PAH compounds as well as oil fractions from the effected environment? Do the physicochemical treatment regimes that we currently use to clean oil spills contain PAH compounds?
Could the factories involved in the pollution of the water supply implement some type of prevention plan that eliminates PAH’s before they reach the environment? If the conditions needed for a specific bacteria, that could eliminate the PAH’s, was found they could introduce the bacteria in a controlled environment before it reaches the outside and is subject to unpredictable conditions.
The filter paper confuses me. Why would they add the paper to the medium? Was the purpose of this to possibly separate larger bacteria from bacteria that could fit through the filter paper?
It says that a phylogenetic tree was drawn based on the 16S rRNA gene sequences but were there any sequences that did not fit? What happens to sequences that are new and how are they placed?
Could the strains be genetically modified to potentially handle higher concentrations of naphthalene?
Could the N16 and N18 strains thrive better in lower levels of naphthalene, or could the environment change the outcomes?
Could some of these strains be used in other bodies of water or are they specific to the Persian Gulf?
Is phenanthrene also located in our oceans? Could they also take samples from water?
Did they only isolate the one phenanthrene – degrading bacterial strain or were there multiple and they used the best one?
Were they looking for a specific morphology or were they just separating them by morphology to make isolation easier?
Were the different substrates added to see which bacteria would grow best with which substrate?
Was the method used here the same method as what we did in lab 7? Initially reading this I did not fully understand what it was saying but after doing it, it makes a lot more sense.
Did they only focus on the results of ZX4 because it was the best at degrading phenanthrene?
Since ZX4 only oxidized 55 of the 95 carbon sources why did they not continue testing with other bacteria that could possibly oxidize the other 40 carbon sources?
Do phenanthrene degrading bacteria always turn yellow or is it just strain ZX4?
What type of genetic modifications would they make to enable the bacteria to degrade PAHs more effectively?
Since the strain was able to degrade other aromatic compounds would this be a good indicator that it should be studied more and used in more areas for clean up?
What made them choose biofilms from the hot springs? Was it based off of the community characterizations?
Could the information discussed in this paper benefit us somehow medically? Could there be some type of use for these bacteria against arsenic poisoning?
What was the reason for the large gap in sampling times? Also they kept samples for several months, how many times would they need to repeat experiments?
What is the purpose of shaking vs not shaking the tubes?
What was the purpose of the great length between collection times? Why did they need to go back the next april?
As we discussed in lab, normally you would expect to find a largely diverse environment full of various species, but with this environment being more harsh there are less species. I find it more interesting that Ectothirohodospira Sp. to be so dominant.
Although other bacterium were found, Ectothiorhodospira sp. was still dominant in the community, like in table one.
I am a little confused about how the chemistry of the spring waters and the shallowness relate? I may be missing something or not understanding completely.
If the dark As(V) and the light As(III) both have optimal activities at the same temperature would this also indicate that a microbe is acting in both light and dark or is it just adaptation by different microbes.
Why did they choose Hydrogen and sulfide as electron donors?
The Ectothiorhodospira sp. have obviously adapted extremely well in this environment. Do you think having the arsenite efflux gene (arsB) added in this adaptation?
PAH is a toxic organic compound which means Polycyclic aromatic hydrocarbons. Microbial degradation studies suggest that PAH is a successful way of removing toxic substances. What are the enzymes that can degrade PAH and what are the different mechanisms that that can degrade PAH? Degradation is usually initiated by hydroxylation. What are the other enzymes that can initiate degradation? What are the different pathways that PAH are transferred? Are these the intermediates? Does anything happen after the central pathways or is that the end result? Why are the aspects of PAH biodegradation unclear? Why isn’t metabolic pathways seen in marine bacteria?
Fluoranthene is a PAH that is composed of a five member ring. Fluoranthene has been used as a model compound for biodegradation because it is similar to other compounds of environment concerns. Only a few fluoranthene have been isolated from the marine environment. Why only a few? Why are there gaps for the mechanisms for fluoranthene degradation? Why is there little information available for marine degraders?
What is involved with complete genome sequencing and how long does it take to get the results.? Has a complete genome sequence been completed? How many PAH degraders been sequenced? What are the results for the Mycobacterium vanbaalenii PYR-1 genome? What is the purpose of using Delftia sp Cs-1. What is the importance of having a genomic island? (to allow for adaptation or something else?) Genomic analyses of Alteromonas sp SN2 metabolisms PAH. What are the 4 pathways for Polaromonas naphthalenivorans CJ2. Research on the mechanism of PAH degradation is not understood. Why? Why are there so many unknown metabolic pathways?
Celeribacter indicus P73T can degrade a lot of PAHs which includes naphthalene and fluoranthene. Strain P73T is a fluoranthene degrading bacterium; It can degrade naphthalene. P73T is gram-negative, rod shaped and non-motile. What are the genes involved with the metabolism of PAH? Why is it only a possible degrading pathway?
that’s interesting. I would rather take the easier route. I don’t have the hardest route all the time. lol So is it impossible to sample other waters? I know you said you would need a boat or a ship. Has it been done before?
I’m not sure this may be in the discussion question
What is the method of Ausubel et al? Why did they isolate samples of P73T from different areas. (P73T from the Indian Ocean and B30 from the Arctic Ocean) What is the significance of 28 degree C? Are they only able to grow at this temp?
What is Solexa paired-end sequencing? Why were there gaps between the assembled P37T scaffolds? Are the gaps necessary for the experiment to work? What is ABI 3730 capillary sequencer? What is Glimmer 3.070? What is island viewer and why was this used to analyze the data?
Why was the gene delection necessary? What is the cre-lox recombination method? Why was E. coli WM3064 used as the donor strain: Why was pJK100 used as the suicide vector? Could another strain be used for the donor strain and the suicide vector?
What was the purpose of using sea water to grown strain P378T? What is GC-MS? What is the chromatographic retention time?
What is Solexa paired-end sequencing? Why were there gaps between the assembled P37T? What is Glimmer 3.070. What toold are available in the IMG server?
that is really interesting. Do you plan to attend graduate school?
I think it is a blood agar plate but I’m not positive on that one
thanks that was one of my questions (what is paired-end sequence) I guess it makes sense because a pair is usually at least two
that was one of my questions. thanks
I think the cre-lox recombination is a knock out process. I googled it and this is what I came up with. In this article they are using mice as a study for cancer therapy. This is an interesting article
I’m not sure. I googled it, I was looking at the wrong results
they seem to use E. coli a lot. I guess they know a lot about it
okay I found what GC-MS means. (Gas chromatography mass spectrometry) and I know what retention time is. It has been awhile for microbiology. I remember now after looking at the definition (the time required for a solute to migrate, or elute, from the column, measured from the instant the sample is injected into the mobile phase stream to the point at which the peak maximum occurs)
The chromosome has the largest bp size when compared to pP73A, pP73B, pP73C, pP73D and pP73E. The chromosome CDS number is higher than the others. In addition, the chromosome is the only one that contains tRNA and sRNA. I noticed that as the size of bp decrease the CDS number decreases, the G + C content decreases, the average CDS size decreases, the codeing density decreases, and CDS assigned to COG decreases.
Why wasn’t a gene found for tRNA-Tyr found; the tyrosyl-tRNA synthetase gene was identified. Could it be that they just couldn’t see it?
This one seems interesting to me because the pP73E is the smallest and doesn’t have a lot of functional categories or cellular processes and signaling. I also noticed that it doesn’t have ribosomes, no cell wall and energy is not produced unless I missed something. Could this be because of the lack of ribosomes or because it is so small? pP73D isn’t much bigger than pP73E but it has some of the things that pP73E does not contain according to this schematic. Does anyone have a different interpretation that would explain why?
What is SIGI-HMM? What is HTG?
why was glucose-6 phosphatase absent?
What is the significance of FADL genes? What does and the proposed fatty acid transporter (FAT) family of proteins suggest?
What is the ATP binding cassette?
What is the function of the plasmid?
what is the function of the plasmids?
What other theories are available besides divergent evolution for ParA and Rep proteins?
What is proteome?
What is NCBI?
What are the other genes that were involved in lipid transport?
Were the genes that encode flagella used for this study? if so, why wasn’t any flagella observed and why was the P73 non-motile? I read that there wasn’t any flagella observed so this would explain why it was non-motile, but it doesn’t make sense if they used the genes that encode flagella.
What other genes were involved in the metabolism of aromatic hydrocarbons?
Did tRNA-Lys and tRNA-Arg transfer genes between each other?
What is protocatechuate? Why were the genes found in the chromosome and in the plasmid? Why were the genes found in the plasmid vs the ones found in the chromosome or genome vs plasmid?
Were test done to determine if these genes actually enhance the catabolic activity of strain P73?
were there test done to determine if these genes enhance the catabolic ability of strain P73?
The reading suggest that BenE may transport benzoate-like aromatic compounds. What evidence supports this theory?
I found the answer to this question: SIGI-HMM is a sequence composition GI prediction method that is part of the Columbo package. This method uses a Hidden Markov Model (HMM) and measures codon usage to identify possible GIs. and HTG I must have missed that it’s in the paper horizontally transferred gene
Oh Ok I was just wandering if there was a reason why it wasn’t there
ATP binding cassette transporters ( ABC transporters) are members of a transport system superfamily that is one of the largest and is possibly one of the oldest families with representatives in all extant phyla from prokaryotes to humans
The main function of plasmids is to carry antibiotic resistant genes and spread them. The other function is to carry those genes which are involved in metabolic activities and they are helpful in digesting the pollutants from the environment
I found the answer to this one: to carry those genes which are involved in metabolic activities and help in digesting the pollutants from the environment. The main function is carry antibiotic resistant genes and spread them
The proteome is the entire set of proteins expressed by a genome, cell, tissue, or organism at a certain time
I’m not sure if this is right: The National Center for Biotechnology Information?
is this sentence it mentions non-redundant protein sequence database…I don’t think that is what NCBI stands for. I will go with my previous statement
thanks. I looked at it
ok…so the region B is the one that may have been involved in the lateral gene transfer?
Fluoranthene degradative pathways seem very complex. Are there plans for anyone to continue this research?
How many of the 138 genes were studied? Were the genes that encode 6 ring hydroxylating dioxygenases and the 8 ring cleaving dioxygenases studied in this experiment? What other catabolic enzymes, transcriptional regulators, and transporters are involved in the degradation pathways?
Because the other dioxygenases can transfer fluoranthene were they not used because they do not belong to the toluene/biphenyl family fluoranthene dioxygenase groupthis an important factor for them to belong to the biphenyl group?
Were we expecting more than 1 strain for P73?
I think I understand this better
I agreee
Because the cost are low and there aren’t any secondary waste treatment this sounds like a win-win situation. However, the high potential for in situ or onsite treatments I’m not to sure about that one. I googled it and it said that the main advantage of in situ treatment is that it allows the soil to be treated without being excavated and transported which sounds like a good idea to me because it said it was cost savings, BUT it said they require longer time periods which could be considered a negative if there as a short deadline
That’s great that 40 species have been isolated from the environment. The bad thing is the article said that little is known about whether PAHs degrading performance can be maintained or encouraged by the presence of heavy metals. I suppose lighter metals will not get the job done and I’m assuming that’s why they used the heavier metals.
I googled ryegrass and it said that ryegrass could take up and transport more ametryn from the soil into the plant shoot. Ametryn is a selective herbicide used for killing weeds. I suppose this would be important if you are using it for hydrocarbon-contaminated soils. Also. ryegrass had the maximum value of translocation factor (TF) for ametryn. I saw a study on google that talked about ryegrass, maize, wheat, and alfalfa. Could they use maize instead?
The three objectives let me know what to look for in the next section. They will test the cooper tolerance, characterize the phylotype and expression of PAH-RHD and c23o genes and study the potential of the remediation of PHE-cooper co-contaminated soils. This paper is easier for me to understand. I suppose because it is the second one. I don’t know. I think I am getting the swing of things
Oh okay I see
Abiotic factors are the non living parts of the environment which include sunlight, temperature, wind, water, soil and naturally occurring events such as storms, fires, and volcanic eruptions
Abiotic factors are the non-living parts of an environment. These include things such as sunlight, temperature, wind, water, soil and naturally occurring events such as storms, fires and volcanic eruptions
MSE with glucose was used as a blank control. I haven’t heard it expressed that way. I’m assuming that it just mean they were used as a control
sorry, my comment on 2 about the blank control was for this paragraph
perhaps water lost or temperature change
Figure 1-B is a phylogenetic tree that shows the common ancestors. Figure 1a appears circular and 1b appears as a rod shape. Sphingomonas is a gram negative aerobic bacteria.
PHE efficiency decreased as mg/L increased as indicated in Fig S1. At 100 mg/L the PHE efficeincy was 100%. At 900 mg/L the PHE efficiency dropped to around 25%
As the mM of copper increased the PHE removal efficiency decreased. At 0 mg of Cu, PHE efficiency was at 100%. At 7.25 mM the Cu mM dropped close to zeo. Previous studies reported a decline in microbial respiration. I this means that cooper stops degradation. The experiment mentioned iron, so does iron help with PAH degradation?
Did they mention the enzymes that were involved in the catabolic pathways? I know that enzymes speed up a reaction
this paragraph is a little confusing to me. So is this the first study that said that copper promote high levels of Cu? That’s what the last sentenced said that’s interesting. I guess they used nickel instead of cooper?
Can they use nickel and copper to increase the growth? (together) This study used copper and if I understand correctly a previous study used nickel.
Rye-grass improved Sphinogobium by proving nutrients. A previous study was done that proved that reyegrass enhances soil peroxidase. Maybe ryegrass can be used to increase other plants growth.
ok that makes sense to me. (Too much of something being toxic) I will just compare this to taking too much medications to keep it straight in my mind
never mind, I think I saw in the other paragraph below
I think it converts both atoms of O2 into a substrate.
yes I did mistype that. I can’t remember what I was thinking but you explained it in class (the graphs) I think I was looking at something on the graphs
I meant providing
It’s not surprising that combustion or oil related anthropogenic activities cause a lot of PAHs. It’s sad because that means that a lot of environmental waste are the cause of these toxins. In a perfect world you would expect the toxins to come from forest fires and natural oil seeps but that is not the case.
I wasn’t good at mechanisms. I need to see the diagram to follow this one. It’s very detailed and I can’t wait to see the diagrams that go with them.
I wander why they used gram negative bacteria. I read somewhere that gram positive worked better for chlorinated dioxins. I’m assuming this one does not use chlorinated dioxins.
a nucleotide sequence is a succession of letters that indicate the order forming allele within a DNA (using GACT) or RNA (GACU) molecule. Sequences are usually presented in the 5′ end to the 3′ end. The sense strand is used for DNA.
Protein exist in different dissociated forms such as amino acids, peptides, enzymes hormones and polypepetides. They carry out different functions in the body. Protein analysis is done to determine the quantity, quality, and the biological reactions in the body. Protein analysis is done to learn the concentration of protein on nutritious food. It is also used to learn the quantity of anemia. Protein analysis is a great way to learn what is going on with a patient. It’s also used in times of crime and forensics examinations. I find this part interesting.
That’s a good question. She said they may not be in the order that they are in the methods section. I think the correct order will be in the results section. Someone correct me if I am wrong
Usually a buffer helps to maintain a constant pH. I don’t know if that what is for in this experiment, but that is what buffers do
good question Lisa. I googled it and I could not find a good definition for this one. I think it has something to do with substrate specificity, but I’m not sure about it either.
In figure 2, it said that horizontal transfer may have occurred. This one is a little different from paper 2 horizontal gene transfer
Southern blot uses DNA and northern uses RNA. I finally have that part understood
In paper 2 Fig 3, P2 and B1 have arrows pointing in both directions. PHE-1 only have arrows pointing to the left. There are large subunits and small units for all of them. There are black and white arrows. In paper 3, the arrows all point to the right. There are also dark gray and light gray arrows which are absent in paper 2. The dark gray arrows indicate the genes involved in the electron transport chain or phthalate degradation. The light gray arrow indicates the genes that are not involved in fluorine oxidation.
No oxidation products were detected. Could this be because they were consumed during the rxn?
I’m not sure about that. They said they were tested but amplification could not be found. I’m thinking that the primers did not work, but then it said they used angular dioxygenase genes (whatever that means)
It also said or fluorene
my confusion here is what is angular dioxygenase. ? I know angular means having angles or sharp corners and dioxygenase is an enzyme that oxidizes a substrate by transferring oxygen from molecular O2 to it , but I couldn’t find a good definition for both words together. Does anybody know?
enzyme biology sounds like an interesting course
good, biochemistry was a bit much for me I would rather deal with organic chemistry and I hated organic
Now the lectures on passive and active transport makes sense. The large ones need transport proteins. This makes sense to me. When they talked about it in lecture I knew what they were talking about, but this helps me form a “mental picture” of it .
Is this what they used to use on our crops? Whatever they are using people are saying that it causes health problems.
so in this one, are they isolating carbofuran to degrade it to make it safer for the environment?
What is the mcd gene? is it malonyl-CoA decarboxylase? or is it something else?
this one is taking about how strain KN65.2 growth conditions. Was E. coli the control again?
Did they use PhyML as a comparsion to strain KN65.2?
Is this similar to isolating DNA or RNA?
exconjugants means a protozoan just after the separation following conjugation. Protozoan is a single cell animal from the group Protista, such as flagellate, ciliate, amoeba or sporozoan.
glucose is the control?
oh! gotcha
I’m not sure what they mean that the rate-determining step is constitutive which means serving to form, compose are make up a unit as a whole. Figure 1 looks almost the same to me except the black circles are slightly lower than the white circles and the mineralized percent increases over time.
the genes are transcribed at a constant level?
it said strain KN65.2 is closely related to the isolate Novosphingobium. I was thinking that they were saying that horizontal gene transfer occurred, but they didn’t mention horizontal gene transfer. I’m assuming that if they thought horizontal gene transfer occurred that they would have mentioned it here.
Only 50% of the wild type was c, so does this mean that carbofuran wasn’t a good choice to degrade the wild-type because only half of the wild type was mineralized?
what I got out of this paragraph is that group IV mutants degraded at a slower rate, very little decomposition, and no growth. Maybe they should try a different gene to see if they obtain the same results
I think this paper was the second most difficult paper. The first was the hardest to understand to me
they used cfd operon. I thought the cfd operon was the reason for the reduced mineralization, so I was thinking for future studies could they use a different gene to obtain more mineralization??
Ok I see that. It said that phthalate it was reported as an intermediate during PAH metabolism
that’s what I understood too.
I get it. I couldn’t see that response at that time
The GC content is good. Strain KN65.2 is similar to the sediment isolate (lateral gene transfer)
I took an economic botany class over the summer & we went over how yeast is not only an important economic and food staple; but also briefly mentioned how it is used experimentally, especially for its enzymatic properties. It’s interesting to be looking into a more detailed account of this.
I find it interesting how PAH-degrading bacteria from that specific location are observed to be more effective. I’m wondering what determines that; environmental factors, bacterial evolution, both, something else?
Seeing the results of PAH degradation will be interesting, I’m expecting to see pretty notable differences between samples.
Is there a reason the prepared slides were coated with gold?
It’s very interesting that other than the toxicity and lack of Nitrogen & Phosphorus, all sites present survivable living conditions including necessary nutrients. Is this perhaps why PAHs are able to thrive and are found in relatively high concentrations?
Although the isolates were able to be characterized by unique traits, they all had the gram negative rod shape in common in order to be most efficient in the division & nutrient uptake processes. I wonder if the reason behind their unique properties can be pinpointed, whether it be from environmental conditions, special differentiation, or something else?
I am now seeing that the PAH degraders belong to different classes of Proteobacteria, which explains their unique characteristics. I am interested to know further details for the reason behind why these specific PAH degraders are observed.
I like the flow of this paper & how unique morphology of isolates were mentioned first, followed by phylogeny which provided further specification, and finally the specific abilities each isolate provides in PAH degradation which can be connected again with their unique appearance.
I am interested to see how much diversity is found in the hydrocarbon-degrading microbial deep sea community based off of the sample and how they are described.
I believe that the hydrocarbon-degrading bacteria communities (which are to be identified) are present within the sediment, closer to the surface, and as vent fluids which have petroleum-chemicals present are moved upwards through the sediment closer the surface the bacterial communities degrade the petroleum.
I am interested to see if the hydrocarbon degrading communities observed are present in both samples, or if each sample contains communities unique to their environment.
The use of phenanthrene to isolate degrading strains is because the mineralization results prior to SIP incubations showed presence of PHE in the sample. Is it because it was the only hydrocarbon found, it was the most abundant, or something else?
From what I gather, in paragraph one they are using the Carbon isotope C14 to find which PAH results in the highest level of
mineralization upon exposure to and incubation with the isotope in order to determine which PAH/sediment sample to conduct further studies upon.
Because of the fact that it couldn’t be shown whether PAH degradation is done wholly by Cycloclasticus or initiated by Cycloclasticus which was then consumed along with the PAH it was degrading by another organism without further study proves the intricacy involved in these types of experiments. It’s so interesting that this initial experiment could tail off in other various directions!
I believe they chose these PAHs for test incubations based off of the known presence of PAHs in the oceanic environments they’re studying from prior research.
The sample is identified as Cycloclasticus which is also found in shallow marine sediments. The fact that similar genes at different loci are more than likely responsible for both species of the genus Cycloclasticus being able to function at different depths/oceanic conditions is very interesting as would be a separate study on their comparison.
Based off of this introductory passage I immediately wonder what adaptations or functions within these microorganisms moderate selenite mineralization in order for it to not reach toxic levels that would harm the microorganism, if it would.
I am interested to see the specifics of the fnr regulator and it’s ability to produce a selenium mineral product. I also wonder if this product is considered waste or if it is recycled somehow?
I find this method of measuring the rate of selenate reduction to be very interesting, I believe it would be the most efficient method. Although I do wonder how accurate the numbers will be due to filtration & ion chromatography processes.
I believe that perhaps antibiotics were added to the medium to ensure growth success and prevent contamination. I don’t think it would have an effect on growth rate, just success.
pECL1e was chosen for further study because it was able to rapidly reduce Se VI, unlike the clone pLAFR3 which was unable to reduce Se VI. The change in nutrient medium color indicates precipitation of Se VI and confirms reductase activity.
I believe that after observing the 4 sub-clones had complete ORF which contained fnr and ogt genes they wanted to see if these genes were necessary for reductase activity to occur. The results of cells containing fnr amplification (the gene they chose to study) formed red colonies on agar with selenate, confirming that fnr is necessary for activation of reductase activity.
I am interested to know which specific genes are required to express FNR & why they aren’t present in E. coli S17-1 despite selenate reductase activity being inherent to E.coli. Is it solely because the lack of oxygen sensing transcription factors?
After reading this I wonder if these proteins are perhaps part of a selenate reductase mechanism in E. coli K-12 since fnr gene is not present. I would be interested to see a further study observing pathways in strain K-12 compared to S17-1 to confirm whether there are multiple reductases utilized under different conditions or completely different pathways are used.
I have seen news articles discussing marine wildlife being killed off because of toxic bacteria in the water. Is fluoranthene a source of this problem? If not, then what is the environmental concern associated with fluoranthene?
Since C. indicus P73T is a naphthalene degrader, is it possible that we could determine other genes similar to this gene which may degrade naphthalene also? or is that unlikely?
This experiment will help them understand the genes of P73T and B30 that may allow degradation of fluoranthene.
How long does this process take? Is it possible that this could be used in our experiment using NP as the only source of carbon? Once the metabolites are determined, will more experimentation have to be done on the metabolites? If so, what experimentation would be necessary?
If I am understanding correctly, Does this mean that the organisms in the order Rhizobiales have a strong ability to degrade PAH? If so, do we have access to any of this bacteria to determine its PAH degradation efficiency?
Since it syas that no closely homologues were found in the P73T genome for the naphthalene family, does that mean that we cannot compare the two or does that only apply to this “dioxygenases” section of the paper?
So would it be helpful for us to look for these genes in our experiment if the genome sequence returns in time?
How could this information be resourceful to us? I am trying to understand our experimentation a little better. Will we use the gas chromatography – mass spectrometry?
Is there any possible experimentation that can be done to explain the source of the gene transfer?
Is it possible that since “P73_0346 is also responsible for the dioxygenation of naphthalene” as stated in paragraph 3, that naphthalene may have the same or very similar degradation pathway?
I looked up bioremediation to find out more about it and it has a high success rate for oil spills. If anyone else is interested in the article, here’s the website:
https://www.sciencedirect.com/topics/earth-and-planetary-sciences/bioremediation
Is it complicated to test the influence of ryegrass planting on the microbial degradation of organic pollutants? If so, why is that? and if that is the reason, does the reactivity of heavy metals and PAHs make it complicated to test for degradation?
Why was MSM supplemented with glucose selected as the control?
I’m missing the purpose of this experiment
Was the bacterium expected to be rod-shaped? Aren’t most bacteria rod shaped? If so, what is the purpose of the transmission electronic microscopy?
An ex situ system is a system which conserves and maintains samples of living organisms outside their natural habitat, in the form of whole plants, seed, pollen, vegetative propagules, tissue or cell cultures.
This information was found on:
https://www.bgci.org/plant-conservation/ex_situ/
I found this interesting because although I knew the PAHs were found in the soil, I did not realize that they were a result of combustion.
So does this mean that the degradative genes were horizontally transferred since the gene is found on a plasmid?
I’m assuming (I may be wrong) by your explanation of the temperature stability of the phosphate buffers that the phosphate buffer is used because of that characteristic. Maybe?
SDS-PAGE:
“Protein separation by SDS-PAGE can be used to estimate relative molecular mass, to determine the relative abundance of major proteins in a sample, and to determine the distribution of proteins among fractions.”
Fohttps://www.ruf.rice.edu/~bioslabs/studies/sds-page/gellab2.htmlund on:
What is the purpose of the derivatization?
Even though information is limited, why couldn’t they still create their own primers to test for gram-negative bacteria?
Cometabolic:
“Cometabolic biodegradation of environmental pollutants is a non-growth linked biological process catalyzed by microorganisms. During this process, microorganisms use non-specific enzymes to degrade environmental pollutants that do not support microbial growth.”
From:
https://www.enviro.wiki/index.php?title=Biodegradation_-_Cometabolic
The information displayed on Table 3 is helpful because it also shows the concentration for Naphthalene, which is low.
It’s interesting that in all of the papers the conclusion is that the derivative genes were acquired by gene transfer.
In the lanes indicating the presence of fluorine, it can be seen that the bands are dark and noticeable. Whereas, the glucose lanes are either light or do not contain any at all.
Why did the effect of copper matter specifically?
Will we do something similar to this in class?
So does the strain lose its ability to degrade once the amount is over 600?
To better understand what carbofuran is, I looked it up to compare it to the PAHs we’ve previously studied and found that it is a toxic ands aromatic pesticide commonly used in agriculture. It’s aromaticity, similar to PAHs, is a reason it is difficult to degrade.
Just to clarify, they believe that this mcd gene is the gene that is capable of degrading the carbofuran. Correct?
Also, they found that Sphingomonads are good at degrading carbofuran. So why is it that they used the Achromobacter sp. which isn’t from the Sphingomonad family?
Strain KN65.2 is the strain of achromobacter, correct? I got confused and thought it was a strain of carbofuran. In the previous paper’s experiments, they were testing the degraders so I assume that they are testing the predicted degrader here also.
Also, Was the purpose of this experimentation just to characterize the strain?
I definitely misinterpreted the part in the introduction where it explains that they captured a strain of Achromobacter. I see here that the strain KN65.2 is a member of the Sphingomonad family.
When the author says no difference was found, is the slight difference between the two not big enough to be identified? Or am i reading the figure wrong? It seems to me that the carbofuran-grown cells mineralized slower than the glucose-grown cells.
It appears to me that strain 3A6 is the most comparable to the wild type strain according to figure 3.
Is it’s degradation level the normal level of copper? Or is copper normally at higher levels than this? Because if it’s normally at higher levels than this, that means that the PHE degradation strain wouldn’t be successful, correct?
I’m interested in why they were not able to obtain an authentic PCR product. Perhaps this will be discussed in the results section. Can I take a wild shot in the dark and guess that the primers did not work as intended?
I did some light reading on the ecosystem of the hot springs. I thought that how the two bacterias the chemolithotrophs and cyanobacteria share a symbiotic relationship to keep their environments ideal for each other.
As(V) is being reduced to AS(III) why would also add 2mM of As(iii) into the assay? Am I missing something fundamental or is this to be sure that As(v) is actually being reduced?
Reading through this section was quite confusing. Are they trying to determine if the bacterial strain is a chemoautotroph or a chemolithotroph? Then above they are looking at a phototrophic bacterial strain also. Did I miss something? This method section is unfriendly and I don’t like it.
It is interesting to see that nitrogen and phosphorus can increase the degradation of PAH. I would like to know how this would work in an event of an oil spill where an increase in nitrogen and phosphorus would lead to an algae bloom. Could there be a way to increase PAH degradation without the use of these nutrients?
I would like to know the time it takes to degrade these compounds is it fast-acting or does it take time to break these compounds down. I would assume it would take longer, due to the nature of aromatic rings being extremely stable. With that said is it possible to introduce this species near waterways where runoffs from agriculture is an issue?
It would be interesting if the same set of genes responsible for PAH degradation in a terrestrial environment is the same as that in the marine environment.
I’m interested in seeing what some of the byproducts these bacteria release in the environment and if we could apply this on a large scale for disaster responses.
Why are they inserting genes into a plasmid? Is it just used as a marker for Kan and Amp resistant colonies?
What is the purpose of extracting the gene of another organism and transforming it into the E.coli?
I was wondering why there is an overexpression of the listed genes? Could the PCR be the possible reason for this?
I am interested in how they are able to catabolize of these large molecules. Do the enzymes that these genes produce target specific sites? Would the enzymes be targeting the ester group? If so what stops them from targeting other organic materials that have ester functionality?
In short the region right after the gene.
That is interesting that Phn A can have a PAH substrate range, but cannot convert monocyclic aromatic hydrocarbons. I’m interested in the mechanism that drives the conversion of substrates.
I was wondering, does understand whether a gene is located on a plasmid or chromosome play a part in gene regulation or is this just trying to understand things from an evolutionary standpoint?
I’m interested in if the beta subunits, sequence features of the protein family are also conserved, like the alpha subunits.
I wonder if they could obtain the same results using the metagenomics approach.
I wonder what approach they would use since the primer product was not obtained. I think the best result would to use transcriptomics.
Since they found other bacteria containing the Rieske fragment within the sample; are these bacteria able to degrade Fla at the same efficiency as Rhodococcus?
ignore my comment above
This table actually does not make any sense to me. If you’re sequencing the Rieske fragment and it is homologous to other organisms, how do you know for sure that what you have in your sample is actually Rhodococcus, without sequencing another area of the DNA that would identify the species?
the reason we use nitrogen is that it makes the narrowest peaks on the graphs. It makes the graphs easier to read and it shows good separation.
I’m interested in the process of extracting the compounds of interest. How does it happen? Could it just be the polarity difference of the chloroform/methanol “pushing” the other compunds out of the bacteria?
I wonder why the degradation of Fla decreases at higher concentrations. Could it be that they are reaching the lethal level of Fla?
What is the relevance of these proteins? Are these the protein responsible for the degradation of Fla?
Why would they list a genus that is not part of the experiment? Is it to show that this motiff is conserved in other genuses? I would rather be more interested in what other species of Rhodococcus has this motiff and whether or not if the other species can degrade Fla just as efficient or the other PAHs as efficiently.
If they used zone formation technique for the Rhodoccus sp. why would they change it to spray plate techniques instead? are they using different plates or because they are using a different substrate that requires this technique?
Since the PAH dioxygenase gene is only 78-bp long and in Rhodococcus sp. 100 bp the gene would be conserved. But what are the other bacterial strains being tested are they from different genus? How does the bp length compare to Rhodococcus sp. within the same genus? Is only a section of this gene sufficient for PAH degradation?
Did they ever do anything with Anaerobranca californiensis?
For the e- donor experiment, what was the purpose of washing the red- biofilm sample?
I was wondering if the environment is in an oxic condition would it not kill the bacteria due to oxygen being toxic?
This would a great time to use proteomics to asses what protein is being produced.
I agree with Sarah! I think that further experimentation would be nice to determine which organism is reducing arsenate.
[Microbial bioremediation, the process of degradation of contaminants by the metabolic activities of microorganisms, is an ecological, economical and safe approach that can be applied to the decontamination of PAHs with minor alteration of the soil (Bamforth and Singleton, 2005).]
The use of microorganisms to resolve issues in the ecosystem has always intrigued me. Instead of searching for the latest technological advance or contraption, simply seeding a contaminated site with a bacteria that is able to reverse the effects of these contaminants by pollutant-degradation is a clever way to resolve this issue.
I also questioned what natural resources Antarctica had to offer. I am surprised to learn that it, indeed, has more to offer than polar bears and ice! Antartica seems to have an already sensitive ecosystem due to its necessarily cold temperatures. In addition to that, the increasing human activity in Antartica has caused a serious threat to the ecosystem. With the global climate at a constant rise, the oil spillages and the storage and utilization of fossil fuels have greatly contributed to the downfall of the Antarctic ecosystem. It is depressing to see the carelessness of humans and how they treat their ecosystem.
The three strains exhibited almost matching growth yields, while S. xenophagum yielded the highest CFU/mL with phenanthrene as the carbon source. The experiment was then repeated using diesel fuel as the energy source, and while S. xenophagum and R. erythropolis showed even more rapid growth results, P. guineae shows no growth yield and was unable to use diesel fuel as an energy source. P. guineae may be more susceptible to the other particles or PAHs that diesel may contain, which may have prohibited its growth. After a quick google search, it is said that diesel contains 40% phenanthrene, which means the 0.2% diesel experiment contain more phenanthrene than the 0.05% phenanthrene experiment. This may be the reason why S. xenophagum and R. erythropolis showed increased growth yields with the diesel.
These Vibrio seem to be the perfect specimen in this experiment. While being free-living, they are also able to grow under identical conditions, making testing much easier and coherent. Any changes made will show a direct effect without any interference between different environments.
Siderophores with the highest affinity for iron are more suited to live in areas where iron levels are low. While some bacteria are incapable of producing siderophore for themselves, they use heterologous siderophores in order to acquire the necessary nutrients. Siderophore-non-producing bacteria, or “cheaters”, are more like “gold-diggers” in my opinion. They use heterologous siderophores in order to meet their necessary needs while without expending further energy required for their synthesis.
I completely agree that the use of varying types of media will allow researchers to further clarify which supplementations will provide the best environment for the observation of V. Fischer and V. harveyi growth. As stated in the previous post, both V. fishery and V. harveyi are able to grow under identical laboratory conditions, making any alterations made easily recognized.
The process of electroporation is incredibly interesting! By inducing a voltage across a cell membrane, it makes it permeable to any introduced genes allowing the cell to uptake any new genes as seen here with the plasmids beings transformed into E. coli. During the pulse of voltage across the membrane, the cell becomes permeable, but after this pulse has stopped, the cell then “closes” with the new genes inside which otherwise would not have been able to permeate the cell.
[These results indicate that the inhibitory substance produced by V. fischeri ES114 accumulates during growth, and moreover, is either regulated by nutrients, that a component present in rich medium masks or destroys the inhibitor, or that V. harveyi can overcome inhibition if the exogenously-supplied culture fluids contain additional nutrients.]
This is why testing with different mediums are necessary to gather the most accurate results possible. V. fischeri is known to produce a siderophore that inhibits the growth V. harveyi. My interpretation of these results show that the environment plays a huge role in how these vibrio species behave and interact with each other. These results provide more answers to how interspecies interactions are either promoting or preventing cohabitation.
[ V. fischeri ES114 releases aerobactin. Thus, when its culture fluids are added to V. harveyi, iron sequestration prevents V. harveyi growth. ]
V. fischeri releases aerobactin which is a bacterial iron chelating siderophore. These siderophores take in and store, or sequester, all the iron in the environment. This prevents the growth of V. harveyi due to the insufficient amounts of iron in the environment caused by V. fischeri’s release of aerobactin. By deleting the first step of aerobactin biosynthesis, iucD, we have found that the fluids from the same V. fischeri, did NOT prevent V. harveyi growth indicating the cause of V. harveyi inhibition is due to the siderophores produced from V. fischeri.
It’s interesting how nature works and balances itself out. If the number of cheaters becomes overwhelming, there will be a lack of iron. As a result, cheaters population will diminish when they reach a certain point as they are constantly taking in public goods and not returning the favor. Their source will diminish, and they will die off due to the lack of their source and the non-cheaters will rebound. It’s like the circle of life.
I completely agree with Makayla. The experiment should be done a few more times in order to prove these results. As with any given data, we want to be sure that it is valid. As aerE is the main reason for aerobactin transport, it is a reasonable explanation to why this is possible.
It is interesting to see the significant amount of total nitrogen loss when there are such a variety of processes that produce nitrogen gas. I understand that nature has its own way of balance itself out; however, under some circumstances this may not always be the case. This a prime example why the loss or lack of one source can cause a chain reaction towards ecosystem function.
I am very interested in seeing the further studies used in order to find which particular organism is perhaps causing this loss of nitrogen in the ecosystem. Seeing that scientists have already found reliable data which indicates similar findings to what they are searching for in different areas shows for significant hope by looking at differences and similarities.
Flow cytometry is being used as a tool to measure the abundance of microbes in the sample taken. It is also used in immunophenotyping as a tool to help diagnose and classify blood cell cancers. It also aids in finding a form of treatment.
This information will definitely help guide researchers on what further procedures can be done in order to further improve results.
In Figure 1, graph B shows how oxygen is depleted as water depth increases. When oxygen is completely depleted, is when we notice an increase in H2S. This is a possible indicator that another source is substituted in as oxygen levels depleted. In the anoxic layer, other sources are used in the absence of oxygen.
I completely agree with your observation of these graphs! In graph a, 14N 15N almost correlates with 15N15N, however in the presence of 15NO2 (b) 15N15N is shown to have completely flatlines while 14N15N proceeded to grow at a stable rate without the competition of 15N15N. It appears that 15N15N is unable to grow when any presence of 14NO2 as seen in graph c.
The coupling of nitrification and anammox proves to be a very sustainable relationship.
At deeper depths, living conditions are definitely less sustainable. There are fewer species that are able to live in habitats with lower oxygen content and little to no light source. These species will also be deprived of many food resources due to lack of species diversity.
I found it interesting that the bacteria found were mono culture where as the Archaea found were more complex. After reading the discussion part it seems that the Archaea are mainly present due to the high volume of salt.
With all the advancements in technology and research, why in 2018 are these harmful compounds still so abundant? Why haven’t more steps been taken before to reduce them? What other ways are there besides bio-remediation to reduce these pollutants?
How long did it take to see the results of bio remediation strategy in The Persian Gulf? Is there any way for these areas to become immune to bio remediation to the point that it becomes ineffective?
Is there a specific reason that these three oil contaminated sites were chosen over others? Why did the samples have to be transported on ice versus at room temperature or at a warmer temperature?
Why did they incubate the culture for 7 days? why not more/less days? Is Gas chromatography the most accurate was to calculate the residual naphthalene? Are there other ways to calculate this amount?
What is the GC-FID method? Are there other methods that would show the same results? Why did the peak of the naphthalene decrease so dramatically?
It’s interesting that of 54 strains only 18 (less than half) actually had an adequate growth rate to be further studied. What caused the other 36 to not have adequate growth?
I wonder when/if there will be research for isolation of the genera as naphthalene-degrader?
It’s interesting that there were no Gram positive bacteria in the study. I wonder what type of pollution/ what source of isolation caused that?
Has this isolation been done in any other waters besides this Gulf? If so I wonder what their results show?
How would the results change if this study was done in an area where oil contamination wasn’t so high?
Are there any other studies that use another cheap and effective method to decontaminate PAHs-contaminated soil that could be compared with this one?
I am curious to see how the results from this paper differ from the first paper we read. Why did they anticipate that this strain would be a good model organism? What characteristics lead to this strain being picked?
From what I found the LB medium is a complex media because the exact chemical composition of every component is unknown. Is that correct?
I looked up strain ZX4 it said it was identified as Sphingomonas paucimobilis. Is that correct?
What other experiments besides shaking flask batch fermentation would show the pH level that the strain would grow well in?
How did they choose the strains that were chosen to construct the phylogeny tree? I see in figure 1 that it says “based on a distance matrix analysis of the 16rDNA sequences”, but what does that mean? How were the four clusters in the phylogeny tree determined?
I looked up more information on Pseudomonas putida and found that it is a Gram-negative, rod shaped, saprotrophic soil bacterium. It produces small colonies or patches. Sphingomonas is also gram negative and is non spore forming chemoheteretrophic, aerobic bacterium. It forms yellow or off white pigmented colonies. I find it interesting how they clustered differently.
I wonder if there are any other reports or if there are and discussions on doing more reports about meta-cleavage operon genes in S. paucimobilis to further prove this study?
Besides the salicyclate pathway is there another way for the Phenanathrene to be metabolized by the strain? Could another study be done to find another way?
Why is PhnH hydrolase necessary for conversion? Without it would anything happen? I attempted to look up more information about it but I didn’t see much about it.
From what I know about Paoha Island, it formed due to several volcanic eruptions. I’m wondering if that is going to contribute to why these organisms are able to gain energy from such high concentrations of As(V) and As(III).
Are there only red-pigmented biofilms in this area? If not what others are there and are studies done on them? If so are the results different/similar?
Why did they choose August, October, and April specifically to collect samples? Also, Why was there a gap, October to April, where no samples were collected?
I’m curious to see the results of the temperature range experiment for the light and dark incubated tubes. How is the temperature difference going to affect the samples? Also, how are the results going to vary due to light versus lack of light.
After examining this figure it seems that As(III) is the electron donor and As(V) is the electron acceptor. The temperature of As(V) light incubated biofilm has a wide temperature span compared to the other two. It also shows that the As(III) light incubated slurries and t he PHS-1 light incubated strains over lap.
Are chemoautotrophic bacteria the only type of bacteria that carry out dissimilatory As(V) reduction? If not, what other types of bacteria are known to carry that process out?
Besides Hydrogen and Sulfide, what else can serve as an electron donor for the chemoautotrophic growth of diverse arsenate-respiring prokaryotes? If there are other electron donors, why weren’t they used?
Could further studies be performed to see which biofilm microbes were associated with the arrA amplicons? If so, what would those studies consist of? Would it help the data set of this study?
Could a study be performed to show this cyclic phenomenon for permanently anaerobic ecosystems? If so, why has it not been done before? Also, what would be considered a permanent anaerobic ecosystem?
[Hydrocarbon contamination in Antarctica has profound effects that have been shown to reshape the structure of microbial communities as well as affecting the abundance of small invertebrate organisms (Saul et al., 2005; Thompson et al., 2007; Powell et al., 2010).]
I understand how the oil spillage affects microbial communities in the sediment, but how does spillage directly affect the small invertebrate organisms? From an article online, I found out that nematodes and copepods are two important invertebrates native to Antartica that are important for the shuffling of organic matter and nutrients.
[ Interestingly, this method is the best choice in the bioremediation of soils with low indigenous PAH-degrading bacteria (Castiglione et al., 2016). ]
The article states in the first paragraph that hydrocarbons introduced into soil communities promotes rearrangement of microbial communities, and then implies here that Antartica has soils with low indigenous PAH-degrading bacteria. Aren’t the newly flourishing microbes feeding off the the hydrocarbons introduced into the soil? Wouldn’t introducing PAH-degrading bacteria still upset the natural balance of indigenous microbial communities? I understand the point of trying to find a bacteria to help reduce hydrocarbon pollution, but I wonder about what effect that will have on natural populations.
Yes, the 16s rRNA gene was sequenced for the three highest metabolizing strains found in the samples. They isolated them from the previously isolated 53 phenanthrene metabolizing bacteria from the original 350. Since the 16s rRNA gene is highly conserved between all prokaryotes, it is a great gene to use for the identification between prokaryotic species. I think the three species listed at the end of the paragraph are the three they isolated.
I was curious about the M9 minimal media, so I researched it. It is a minimal, salts based medium to use when you want to grow bacteria with a controlled carbon source and other controlled substances. This gives you the ability to control the carbon source to hopefully isolate certain bacterial colonies; however, the nitrogen source is fixed as it has ammonia present in it.
Although the results clearly support S. xenophagum (D43FB) as being the most efficient PAH-degrading bacteria isolated in the samples, I still have my doubts. For starters, there’s the issue of high cadmium levels (see comment above). Additionally, not stated in the article is the issue of the optimal growth temperatures needed for the strain. The strain grows optimally around 28 degrees Celsius, with negligible growth at 4 degrees Celsius. From googling, I found the average temperature in Antartica to be -10 degrees Celsius on the coasts, and -60 degrees Celsius more inland. How well will this species degrade phenanthrene when actually used for bioremediation, and will a small benefit outweigh the potential effects of artificial seeding?
The article states that the diesel-exposed soil sites in Antartica have levels of cadmium between 15-85 mg/Kg, and the D43FB strain can only metabolize around 20% of the initial phenanthrene when in the presence of high cadmium levels. Although the D43FB strain seems to be the best option due to highest levels of phenanthrene/diesel fuel degradation and biofilm/adhesion formations to phenanthrene crystals, how is the metabolism of phenanthrene by the other two strains affected in response to high cadmium levels?
I think the researchers are just saying that the techniques they used may have favored different bacterial strains than ones that would actually grow in the soil’s native conditions. They achieved their goal of isolating PAH degraders, but there may be ones not detected in the laboratory conditions that readily grow in the soil. They are simply stating a limitation to their techniques.
I could be wrong, but I would assume the bacteria would have a greater chance of forming biofilms in the natural environment compared to the laboratory. I think this because bacteria sometimes form biofilms so that they thrive better in harsh environments and can work together to become more resistant to certain conditions. I think attempting to simulate the soil’s conditions would tell us a lot more, especially considering how the D43FB strain had genes encoding for flagellar components, but chemotaxis was not observed in laboratory conditions. This was important in showing how bacteria can turn on/off certain genes according to their environments.
I could not find compounds similar to siderophores, but I did find that some organisms are able to store excess phosphorous inside of the cell by making polyphosphates. Certain anaerobic bacteria are able to consume carbon energy sources by using these polyphosphates as energy rather than oxygen as an electron acceptor.
From researching, I found that there are hundreds of different siderophores that can have different affinities to iron based on their specific properties. They contain two negative oxygen molecules that bind to the the positive ferric iron, and come in the form of hydroxamate, catecholate, and carboxylate functional groups. The oxygen molecules can be replaced with a nitrogen or sulfur molecule, but this decreases affinity. Additionally, the functional groups can act as hexdentate, tetradentate, or bidentate ligands. They go from the highest affinity to lowest affinity for iron in that order.
I was curious about the CAS assay so decided to research it. Chrome azurol S (CAS) and hexadecyltrimethylammonium bromide (HDTMA) are used to make a complex that binds ferric iron. It can be used to quantify siderophore levels in a sample because as siderophores remove ferric iron from the complex, the absorbance of the fluid sample will change. This assay cannot be used to tell you which siderophores are present because since there are hundreds of siderophores with different binding affinities, this would require several assays that are more specific for certain siderophores. The absorbance of the fluid should decrease when more siderophores are present.
I am a little confused with this sentence. Polymyxin B acts as an antibiotic to gram-negative bacteria because it binds to the lipopolysaccharides of the outer membrane causing the permeability of the membrane to change, leading to cell death. Did the antibiotic specifically only target the V. fischeri because the V. harveyi were conjugated with an antibiotic resistance plasmid from treated E. coli? This is what I assume happened based on the first paragraph of the paper, but was a bit confused on that as well.
The takeaway from this paragraph and figure three is that the Aerobactin siderophore produced by V. Fischeri is controlled by a biosynthetic pathway, since it shows negative control of a repressible operon. Under normal conditions when iron is present in sufficient amounts, it acts as a corepressor and binds Fur, which binds DNA and stops the transcription of siderophores. When iron is depleted, the Fur repressor is not active and transcription of aerobactin is turned on.
Figure 3 proves the Fur gene to be responsible for repressing siderophore production. Figure 3A uses a CAS assay to show that as iron concentration increases, siderophore production decreases; this is not seen in the mutant with the removed Fur gene. In Figure 3B, the promoter for the target gene was coupled with mVenus, a fluorescent protein that can be quantified. As iron increased, mVenus was picked up less, meaning that transcription was repressed as iron increased. Again, there is no effect seen on the mutant.
I believe that this was just an example of what the authors stated earlier in the paragraph. It is a cheater strain that is able to utilize various siderophores secreted by other strains but does not incur any costs by producing it’s own. I could not find on the internet whether it lost it’s biosynthetic genes for producing it’s siderophore or simply gained receptor and importer genes.
The aerE gene is still in the cheater strains because they will still need to export the siderophore out of the cytoplasm. Having this gene is beneficial because it allows the siderophore to not build up to toxic levels intracellularly, and it also allows the cheater to cycle the siderophore for more iron. Because of these advantages, these strains are probably selected for at a higher degree since they increase the cheater’s fitness.
Upwellings make ammonia available to bacteria, who can oxidize this to nitrate for energy. This creates areas of low oxygen, so bacteria have to use nitrate or nitrite as terminal electron acceptors. When they do this, they use either denitrification or anammox for energy. Denitrification involves the reduction of nitrate to nitrogen gas. Bacteria only switch to this in anoxic conditions when no ammonia is present because this reduction consumes energy. Anammox involves oxidizing nitrate to nitrogen gas by using nitrite as the terminal electron acceptor when no oxygen is present.
I was curious about the anoxic conditions of the Black Sea, and found that this is mostly due to there being two layers of water in the sea. Rainwater and river water flows into the sea causing the surface of the sea to be composed of freshwater, while the deeper waters are fed by the Aegean Sea and are much saltier. This creates a large density difference, so the two layers do not mix well. This creates highly anoxic conditions in the deeper layers of the sea.
PCR techniques will allow the researchers to quantify the expression of the amoA gene in different nitrifiers. Additionally, PCR of the 16S rRNA gene and phylogenetic analyses will allow for identification and characterization of the bacteria/archaea that they find.
Since 14N is the most abundant isotope in nature, 15N can be added in so that the researchers can use this ratio to determine the uptake and usage of 15N. During anammox, bacteria use ammonia and nitrite to produce nitrogen gas and water. Aerobic ammonia oxidizing bacteria can convert ammonia to nitrite and nitrogen gas; therefore, the aerobic oxidizers produce a nitrite product that the anammox bacteria can use. The researchers will be able to study exactly how this nitrogen is shuffled around between the two by using 15N.
In the suboxic depth, nitrate levels drop as ammonium levels rise. Anammox bacteria also peak in this area as they oxidize ammonia to nitrogen gas using nitrite since there is no oxygen.
I believe that this figure is simulating the conditions of the different depths by making different nitrogen species available. Only (a) shows production of nitrogen gas made fully of 15N when ammonia is available. When NO2 is available (b and c), this is not produced. This means that in the suboxic and anoxic zones where ammonia is being oxidized by anammox bacteria, ammonia is being used and converted to nitrite and nitrate in order to fuel the anammox. When nitrite is already available, this doesn’t happen.
These results state what I interpreted from the graph; that in the suboxic zones where only ammonia is present, anammox is linked to nitrification to replenish nitrite.
I think the conclusions the authors draw are very interesting. Due to anammox rich areas being shown to be linked to nitrification, this gives a more clear picture into marine nitrogen loss. It would be very interesting to replicate these types of experiments in the oceanic OMZs.